Structural insights into Semiliki forest virus receptor binding modes indicate novel mechanism of virus endocytosis.
Yang, D., Wang, N., Du, B., Sun, Z., Wang, S., He, X., Wang, J., Zheng, T., Chen, Y., Wang, X., Wang, J.(2024) PLoS Pathog 20: e1012770-e1012770
- PubMed: 39705215
- DOI: https://doi.org/10.1371/journal.ppat.1012770
- Primary Citation of Related Structures:
8YVY, 8YVZ, 8YW0, 8YW1, 8YW2 - PubMed Abstract:
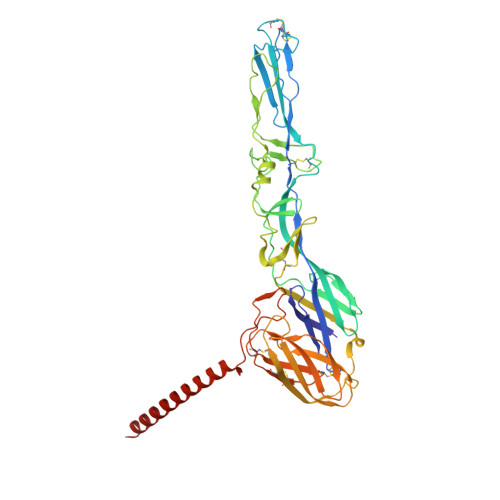
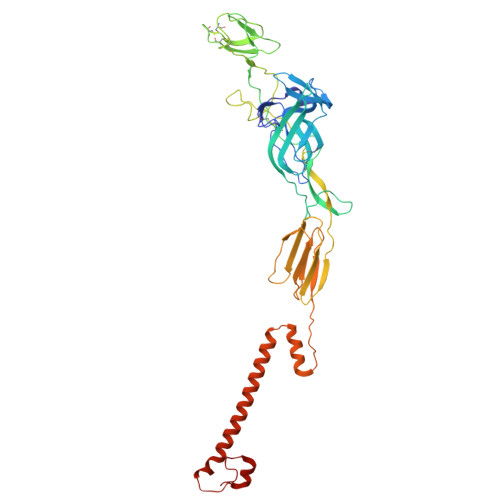
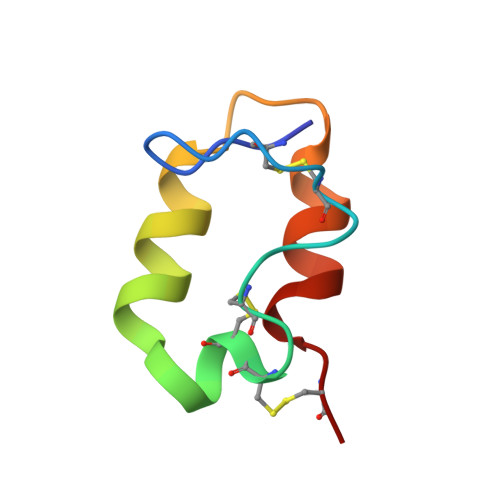
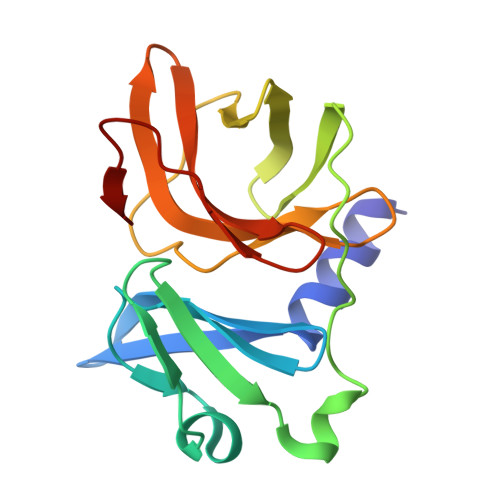
The Very Low-Density Lipoprotein Receptor (VLDLR) is an entry receptor for the prototypic alphavirus Semliki Forest Virus (SFV). However, the precise mechanisms underlying the entry of SFV into cells mediated by VLDLR remain unclear. In this study, we found that of the eight class A (LA) repeats of the VLDLR, only LA2, LA3, and LA5 specifically bind to the native SFV virion while synergistically promoting SFV cell attachment and entry. Furthermore, the multiple cryo-electron microscopy structures of VLDLR-SFV complexes and mutagenesis studies have demonstrated that under physiological conditions, VLDLR primarily binds to E1-DIII of site-1, site-2, and site-1' at the twofold symmetry axes of SFV virion through LA2, LA3, and LA5, respectively. These findings unveil a novel mechanism for viral entry mediated by receptors, suggesting that conformational transitions in VLDLR induced by multivalent binding of LAs facilitate cellular internalization of SFV, with significant implications for the design of antiviral therapeutics.
- State Key Laboratory for Animal Disease Control and Prevention & National Data Center for Animal Infectious Diseases, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, People's Republic of China.
Organizational Affiliation: