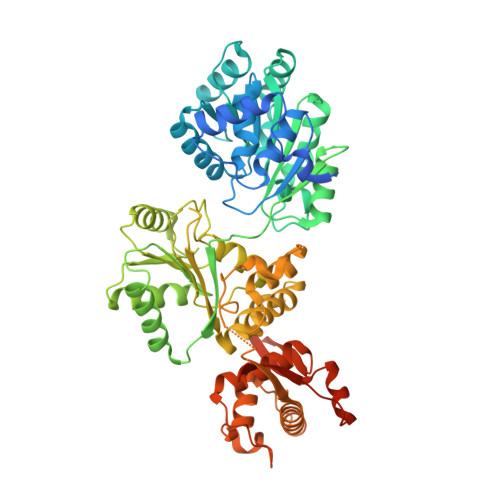
Efficient coordination between the winged helix domain and the aromatic-rich loop restructures the ATPase domain and facilitates DNA unwinding by human RECQ1.
Das, T., Mukhopadhyay, S., Das, A.K., Ganguly, A.(2025) Nucleic Acids Res 53
- PubMed: 40512545
- DOI: https://doi.org/10.1093/nar/gkaf489
- Primary Citation of Related Structures:
8YRS - PubMed Abstract:
RecQ helicases can unwind a wide spectrum of DNA structures and thereby protect the cells from genome instability. Unwinding mechanisms have been extensively studied for bacterial and human RecQ helicases. DNA-induced winged helix (WH) domain repositioning and allosteric remodeling of the ATPase domain have been shown to be important for unwinding activity of bacterial RecQ helicases. In contrast, no such altered conformational state was observed for human RECQ1 upon DNA or nucleotide binding. In this study, we have crystallized and characterized an engineered RECQ1 containing a flexible glycine serine-rich linker inserted between the zinc binding and WH domains. The linker containing construct exhibits more efficient DNA binding and unwinding activity. A crystal structure of the engineered RECQ1 in complex with DNA exhibits conformational rearrangements of the helicase and WH domains, resulting in a more compact structure. Our structure, for the first time, demonstrates that alteration of the distance between the tip of the β-hairpin and the ARL favors DNA binding and remodels the ATPase domain, leading to alteration in substrate recognition and unwinding activity. These structural rearrangements are necessary for efficient coordination between the WH domain and the helicase domain, coupling DNA binding and ATP hydrolysis to strand separation.
- Department of Bioscience and Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur 721302, West Bengal, India.
Organizational Affiliation: