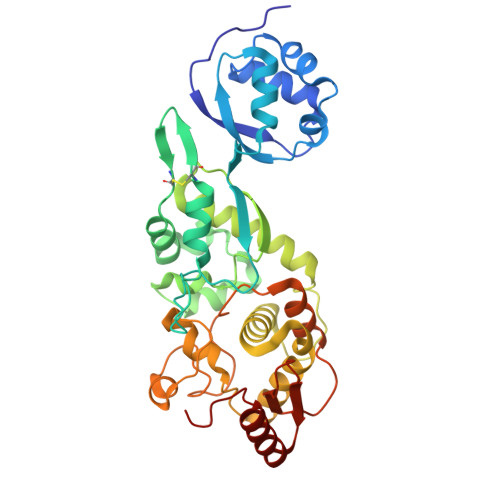
Structure of MltG from Mycobacterium abscessus reveals structural plasticity between composed domains.
Lee, G.H., Kim, S., Kim, D.Y., Han, J.H., Lee, S.Y., Lee, J.H., Lee, C.S., Park, H.H.(2024) IUCrJ 11: 903-909
- PubMed: 39297240
- DOI: https://doi.org/10.1107/S2052252524008443
- Primary Citation of Related Structures:
8YOA - PubMed Abstract:
MltG, a membrane-bound lytic transglycosylase, has roles in terminating glycan polymerization in peptidoglycan and incorporating glycan chains into the cell wall, making it significant in bacterial cell-wall biosynthesis and remodeling. This study provides the first reported MltG structure from Mycobacterium abscessus (maMltG), a superbug that has high antibiotic resistance. Our structural and biochemical analyses revealed that MltG has a flexible peptidoglycan-binding domain and exists as a monomer in solution. Further, the putative active site of maMltG was disclosed using structural analysis and sequence comparison. Overall, this study contributes to our understanding of the transglycosylation reaction of the MltG family, aiding the design of next-generation antibiotics targeting M. abscessus.
- College of Pharmacy, Chung-Ang University, Seoul 06974, Republic of Korea.
Organizational Affiliation: