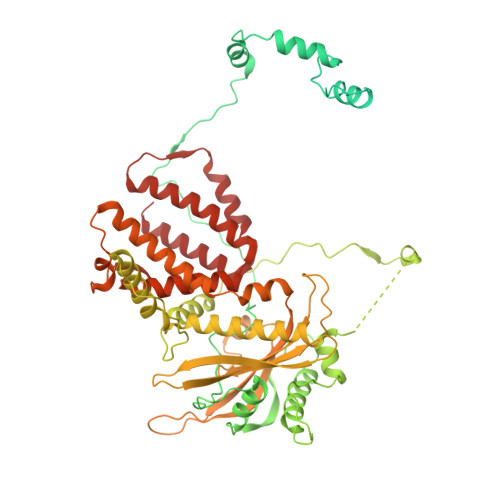
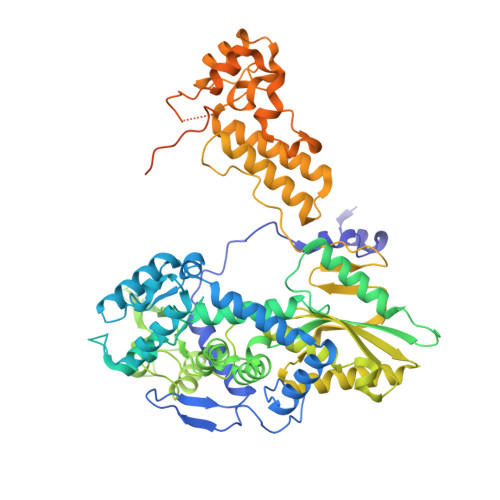
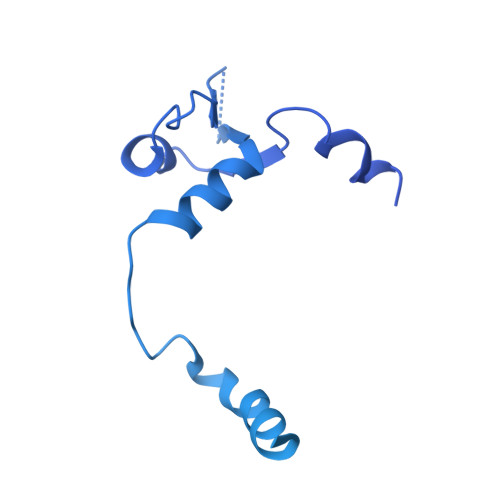
NS2 induces an influenza A RNA polymerase hexamer and acts as a transcription to replication switch.
Sun, J., Kuai, L., Zhang, L., Xie, Y., Zhang, Y., Li, Y., Peng, Q., Shao, Y., Yang, Q., Tian, W.X., Zhu, J., Qi, J., Shi, Y., Deng, T., Gao, G.F.(2024) EMBO Rep 25: 4708-4727
- PubMed: 39026012
- DOI: https://doi.org/10.1038/s44319-024-00208-4
- Primary Citation of Related Structures:
8Y7M, 8Y7O - PubMed Abstract:
Genome transcription and replication of influenza A virus (FluA), catalyzed by viral RNA polymerase (FluAPol), are delicately controlled across the virus life cycle. A switch from transcription to replication occurring at later stage of an infection is critical for progeny virion production and viral non-structural protein NS2 has been implicated in regulating the switch. However, the underlying regulatory mechanisms and the structure of NS2 remained elusive for years. Here, we determine the cryo-EM structure of the FluAPol-NS2 complex at ~3.0 Å resolution. Surprisingly, three domain-swapped NS2 dimers arrange three symmetrical FluPol dimers into a highly ordered barrel-like hexamer. Further structural and functional analyses demonstrate that NS2 binding not only hampers the interaction between FluAPol and the Pol II CTD because of steric conflicts, but also impairs FluAPol transcriptase activity by stalling it in the replicase conformation. Moreover, this is the first visualization of the full-length NS2 structure. Our findings uncover key molecular mechanisms of the FluA transcription-replication switch and have implications for the development of antivirals.
- College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, 030801, China.
Organizational Affiliation: