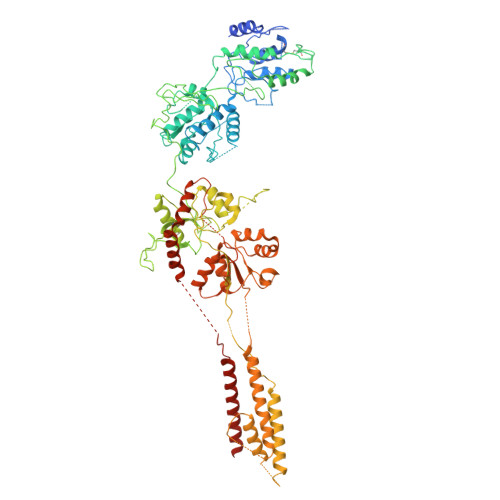
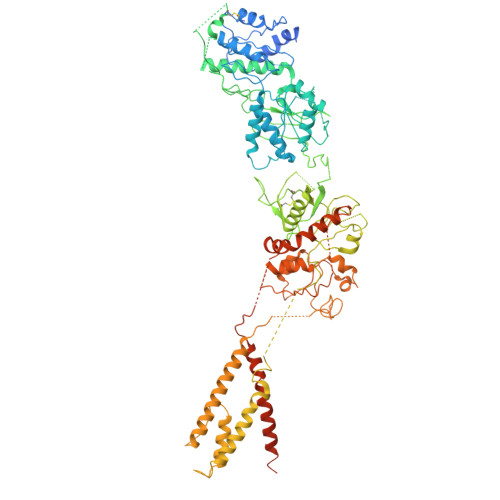
An antidepressant mechanism underlying the allosteric inhibition of GluN2D-incorporated NMDA receptors at GABAergic interneurons.
Zhang, J., Duan, J., Li, W., Wang, X., Ren, S., Ye, L., Liu, F., Tian, X., Xie, Y., Huang, Y., Sun, Y., Song, N., Li, T., Cai, X., Liu, Z., Zhou, H., Huang, C., Li, Y., Zhu, S., Guo, F.(2025) Sci Adv 11: eadq0444-eadq0444
- PubMed: 40043126
- DOI: https://doi.org/10.1126/sciadv.adq0444
- Primary Citation of Related Structures:
8Y1V - PubMed Abstract:
N -methyl-d-aspartate receptors (NMDARs), key excitatory ion channels, have gained attention as anti-depression targets. NMDARs consist of two GluN1 and two GluN2 subunits (2A-2D), which determine their pharmacological properties. Few compounds selectively targeting GluN2 subunits with antidepressant effects have been identified. Here, we present YY-23, a compound that selectively inhibits GluN2C- or GluN2D-containing NMDARs. Cryo-EM analysis revealed that YY-23 binds to the transmembrane domain of the GluN2D subunit. YY-23 primarily affects GluN2D-containing NMDARs on GABAergic interneurons in the prefrontal cortex, suppressing GABAergic neurotransmission and enhancing excitatory transmission. Behavioral assays demonstrate YY-23's rapid antidepressant effects in both stress-naïve and stress-exposed models, which are lost in mice with global or selective knockout of the grin2d gene in parvalbumin-positive interneurons. These findings highlight GluN2D-containing NMDARs on GABAergic interneurons as potential depression treatment targets.
- Institute of Neuroscience, CAS Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: