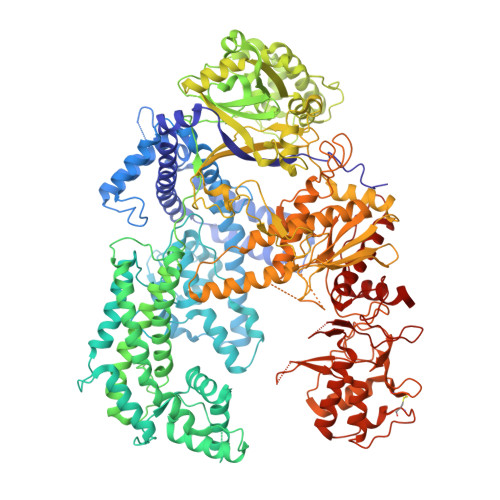
DNA target binding-induced pre-crRNA processing in type II and V CRISPR-Cas systems.
Chen, J., Lin, X., Xiang, W., Chen, Y., Zhao, Y., Huang, L., Liu, L.(2025) Nucleic Acids Res 53
- PubMed: 39676682
- DOI: https://doi.org/10.1093/nar/gkae1241
- Primary Citation of Related Structures:
8Y03, 8Y04, 8Y05, 8Y06, 8Y07, 8Y08, 8Y09, 8Y0A, 8Y0B, 8Y0C, 8Y0D - PubMed Abstract:
Precursor (pre)-CRISPR RNA (crRNA) processing can occur in both the repeat and spacer regions, leading to the removal of specific segments from the repeat and spacer sequences, thereby facilitating crRNA maturation. The processing of pre-crRNA repeat by Cas effector and ribonuclease has been observed in CRISPR-Cas9 and CRISPR-Cas12a systems. However, no evidence of pre-crRNA spacer cleavage by any enzyme has been reported in these systems. In this study, we demonstrate that DNA target binding triggers efficient cleavage of pre-crRNA spacers by type II and V Cas effectors such as Cas12a, Cas12b, Cas12i, Cas12j and Cas9. We show that the pre-crRNA spacer cleavage catalyzed by Cas12a and Cas9 has distinct characteristics. Activation of the cleavage activity in Cas12a is induced by both single-stranded DNA (ssDNA) and double-stranded DNA target binding, whereas only ssDNA target binding triggers cleavage in Cas9 toward the pre-crRNA spacer. We present a series of structures elucidating the underlying mechanisms governing conformational activation in both Cas12a and Cas9. Furthermore, leveraging the trans-cutting activity of the pre-crRNA spacer, we develop a one-step DNA detection method characterized by its simplicity, high sensitivity, and excellent specificity.
- State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Faculty of Medicine and Life Sciences, Xiamen University, No. 4221, Xiang'an South Road, Xiamen 361102, China.
Organizational Affiliation: