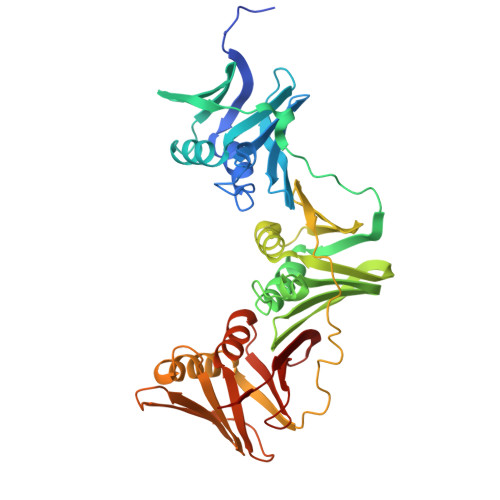
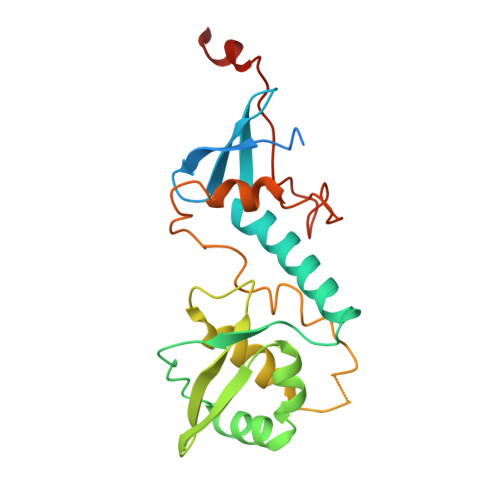
The structure of the MutL-CTD:processivity-clamp complex provides insight regarding strand discrimination in non-methyl-directed DNA mismatch repair.
Nirwal, S., Jha, R., Narayanan, N., Sharma, M., Kulkarni, D.S., Sharma, D., Babu, A.S., Suthar, D.K., Rao, D.N., Nair, D.T.(2025) Nucleic Acids Res 53
- PubMed: 39988319
- DOI: https://doi.org/10.1093/nar/gkaf094
- Primary Citation of Related Structures:
8XLA - PubMed Abstract:
Many prokaryotes, including members of the Neisseria species, lack MutH and cannot employ methyl-directed DNA mismatch repair (MMR). The nick on the daughter strand is created by the endonuclease activity present in the C-terminal domain (CTD) of the MutL homodimer. MutL-CTD is known to interact with the processivity-clamp. The crystal structure of the homodimeric MutL-CTD from Neisseria (NgoL-CTD) in complex with homodimeric processivity-clamp (Nβ-Clamp) shows that each NgoL-CTD monomer binds to a Nβ-Clamp monomer through the conserved motif III (517QHLLIP522). The structure and allied biochemical studies plus in vivo growth assays conducted with wild-type (wt) plus mutant proteins shows that the endonuclease dimer sits transversely across the C-terminal face of the Nβ-Clamp ring. The comparison of the structure with that of the partial prokaryotic replisome suggests that the relative orientation of DNA, Nβ-Clamp, and NgoL-CTD may direct the daughter strand towards one of the active sites in endonuclease homodimer. Nicking assays conducted with wt and mutant NgoL-CTD in the presence and absence of Nβ-Clamp support this inference. Overall, our studies posit that strand discrimination in non-methyl-directed MMR is achieved through a structural strategy involving the β-Clamp which is distinct from the chemical strategy employed in prokaryotes like Escherichia coli.
- Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad 121001, Haryana (NCR Delhi), India.
Organizational Affiliation: