Structural Insights into the Rrp4 Subunit from the Crystal Structure of the Thermoplasma acidophilum Exosome.
Park, S., Kim, H.S., Bang, K., Han, A., Shin, B., Seo, M., Kim, S., Hwang, K.Y.(2024) Biomolecules 14
- PubMed: 38927025
- DOI: https://doi.org/10.3390/biom14060621
- Primary Citation of Related Structures:
8XFX, 8XIE - PubMed Abstract:
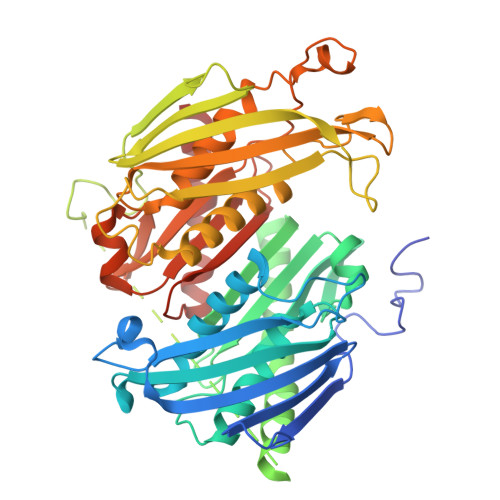
The exosome multiprotein complex plays a critical role in RNA processing and degradation. This system governs the regulation of mRNA quality, degradation in the cytoplasm, the processing of short noncoding RNA, and the breakdown of RNA fragments. We determined two crystal structures of exosome components from Thermoplasma acidophilum ( Taci ): one with a resolution of 2.3 Å that reveals the central components ( Taci Rrp41 and Taci Rrp42), and another with a resolution of 3.5 Å that displays the whole exosome ( Taci Rrp41, Taci Rrp42, and Taci Rrp4). The fundamental exosome structure revealed the presence of a heterodimeric complex consisting of Taci Rrp41 and Taci Rrp42. The structure comprises nine subunits, with Taci Rrp41 and Taci Rrp42 arranged in a circular configuration, while Taci Rrp4 is located at the apex. The RNA degradation capabilities of the Taci Rrp4:41:42 complex were verified by RNA degradation assays, consistent with prior findings in other archaeal exosomes. The resemblance between archaeal exosomes and bacterial PNPase suggests a common mechanism for RNA degradation. Despite sharing comparable topologies, the surface charge distributions of Taci Rrp4 and other archaea structures are surprisingly distinct. Different RNA breakdown substrates may be responsible for this variation. These newfound structural findings enhance our comprehension of RNA processing and degradation in biological systems.
- Department of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul 02841, Republic of Korea.
Organizational Affiliation: