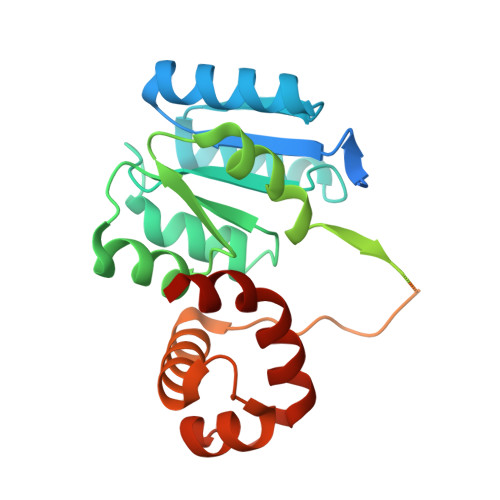
The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Zhou, C.M., Li, J.X., Zhang, T.Q., Xu, Z.G., Ma, M.L., Zhang, P., Wang, J.W.(2024) Proc Natl Acad Sci U S A 121: e2319335121-e2319335121
- PubMed: 38198526
- DOI: https://doi.org/10.1073/pnas.2319335121
- Primary Citation of Related Structures:
8XAS, 8XAT - PubMed Abstract:
The phytohormone cytokinin has various roles in plant development, including meristem maintenance, vascular differentiation, leaf senescence, and regeneration. Prior investigations have revealed that cytokinin acts via a phosphorelay similar to the two-component system by which bacteria sense and respond to external stimuli. The eventual targets of this phosphorelay are type-B ARABIDOPSIS RESPONSE REGULATORS (B-ARRs), containing the conserved N-terminal receiver domain (RD), middle DNA binding domain (DBD), and C-terminal transactivation domain. While it has been established for two decades that the phosphoryl transfer from a specific histidyl residue in ARABIDOPSIS HIS PHOSPHOTRANSFER PROTEINS (AHPs) to an aspartyl residue in the RD of B-ARRs results in a rapid transcriptional response to cytokinin, the underlying molecular basis remains unclear. In this work, we determine the crystal structures of the RD-DBD of ARR1 (ARR1 RD-DBD ) as well as the ARR1 DBD -DNA complex from Arabidopsis . Analyses of the ARR1 DBD -DNA complex have revealed the structural basis for sequence-specific recognition of the GAT trinucleotide by ARR1. In particular, comparing the ARR1 RD-DBD and ARR1 DBD -DNA structures reveals that unphosphorylated ARR1 RD-DBD exists in a closed conformation with extensive contacts between the RD and DBD. In vitro and vivo functional assays have further suggested that phosphorylation of the RD weakens its interaction with DBD, subsequently permits the DNA binding capacity of DBD, and promotes the transcriptional activity of ARR1. Our findings thus provide mechanistic insights into phosphorelay activation of gene transcription in response to cytokinin.
- National Key Laboratory of Plant Molecular Genetics, Chinese Academy of Sciences Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Organizational Affiliation: