crystal structure of the CobH-like domain in cobalt regulator CoaR
Xichun, L.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mercuric resistance operon regulatory protein precorrin isomerase | 218 | Synechocystis sp. PCC 6803 substr. Kazusa | Mutation(s): 0 Gene Names: merR | 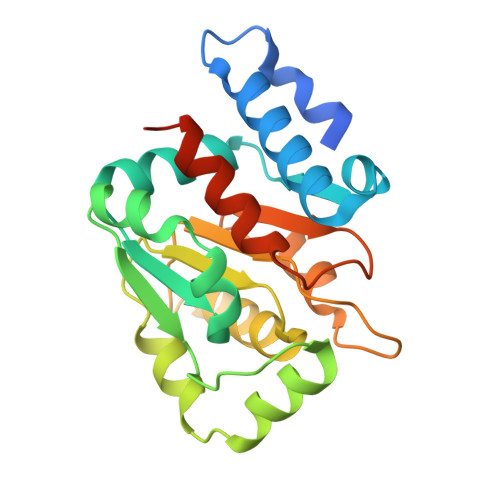 | |
UniProt | |||||
Find proteins for Q55938 (Synechocystis sp. (strain ATCC 27184 / PCC 6803 / Kazusa)) Explore Q55938 Go to UniProtKB: Q55938 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q55938 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL Query on CL | C [auth A] D [auth A] E [auth A] F [auth B] G [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.236 | α = 90 |
| b = 96.236 | β = 90 |
| c = 148.889 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| autoPROC | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, China) | China | 22107046 |