Cryo-EM structure of human DRA (SLC26A3) bound with oxalate
Yang, X., Zhang, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chloride anion exchanger | 775 | Homo sapiens | Mutation(s): 0 Gene Names: SLC26A3 Membrane Entity: Yes | 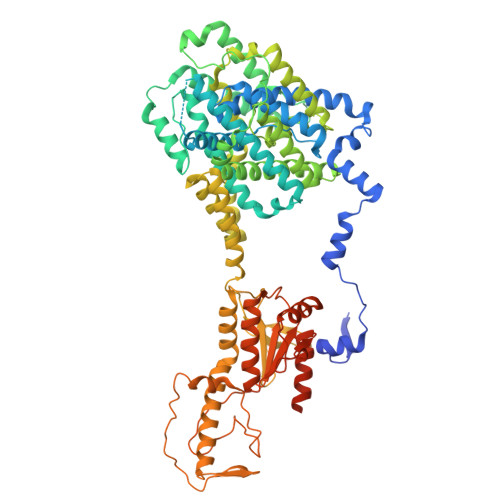 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P40879 (Homo sapiens) Explore P40879 Go to UniProtKB: P40879 | |||||
PHAROS: P40879 GTEx: ENSG00000091138 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P40879 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| POV Query on POV | D [auth A], GC [auth B], HC [auth B], PA [auth A] | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate C42 H82 N O8 P WTJKGGKOPKCXLL-PFDVCBLKSA-N |  | ||
| LPE Query on LPE | AA [auth A] DB [auth B] EC [auth B] FB [auth B] G [auth A] | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE C26 H57 N O6 P XKBJVQHMEXMFDZ-AREMUKBSSA-O |  | ||
| CLR (Subject of Investigation/LOI) Query on CLR | MB [auth B], RA [auth A] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| C14 Query on C14 | AC [auth B] E [auth A] I [auth A] KA [auth A] OB [auth B] | TETRADECANE C14 H30 BGHCVCJVXZWKCC-UHFFFAOYSA-N |  | ||
| D12 Query on D12 | BA [auth A] BC [auth B] F [auth A] FC [auth B] HA [auth A] | DODECANE C12 H26 SNRUBQQJIBEYMU-UHFFFAOYSA-N |  | ||
| D10 Query on D10 | AB [auth B] BB [auth B] CA [auth A] CB [auth B] CC [auth B] | DECANE C10 H22 DIOQZVSQGTUSAI-UHFFFAOYSA-N |  | ||
| OXL (Subject of Investigation/LOI) Query on OXL | C [auth A], SA [auth B] | OXALATE ION C2 O4 MUBZPKHOEPUJKR-UHFFFAOYSA-L |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |