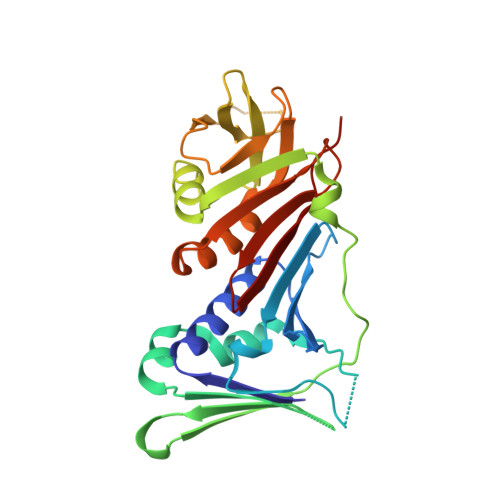
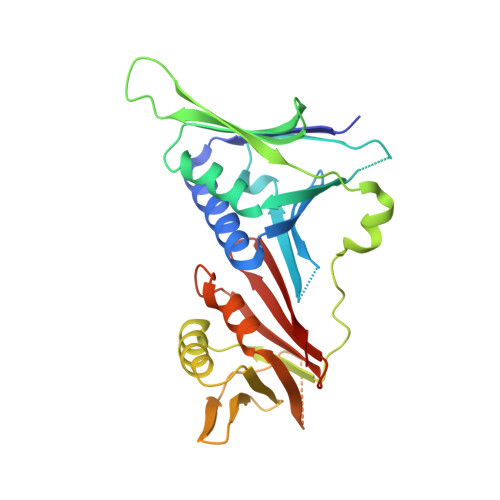
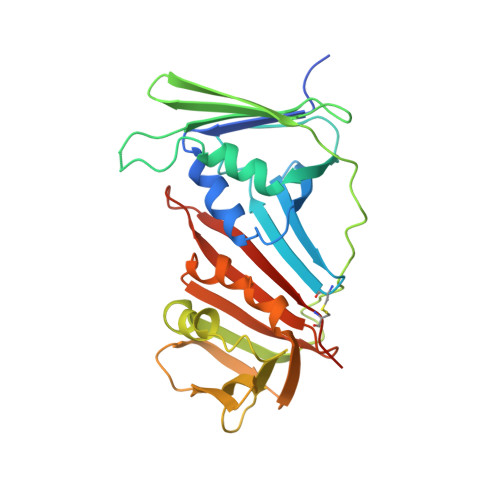
Structural basis for intra- and intermolecular interactions on RAD9 subunit of 9-1-1 checkpoint clamp implies functional 9-1-1 regulation by RHINO.
Hara, K., Tatsukawa, K., Nagata, K., Iida, N., Hishiki, A., Ohashi, E., Hashimoto, H.(2024) J Biological Chem 300: 105751-105751
- PubMed: 38354779
- DOI: https://doi.org/10.1016/j.jbc.2024.105751
- Primary Citation of Related Structures:
8WU8 - PubMed Abstract:
Eukaryotic DNA clamp is a trimeric protein featuring a toroidal ring structure that binds DNA on the inside of the ring and multiple proteins involved in DNA transactions on the outside. Eukaryotes have two types of DNA clamps: the replication clamp PCNA and the checkpoint clamp RAD9-RAD1-HUS1 (9-1-1). 9-1-1 activates the ATR-CHK1 pathway in DNA damage checkpoint, regulating cell cycle progression. Structure of 9-1-1 consists of two moieties: a hetero-trimeric ring formed by PCNA-like domains of three subunits and an intrinsically disordered C-terminal region of the RAD9 subunit, called RAD9 C-tail. The RAD9 C-tail interacts with the 9-1-1 ring and disrupts the interaction between 9-1-1 and DNA, suggesting a negative regulatory role for this intramolecular interaction. In contrast, RHINO, a 9-1-1 binding protein, interacts with both RAD1 and RAD9 subunits, positively regulating checkpoint activation by 9-1-1. This study presents a biochemical and structural analysis of intra- and inter-molecular interactions on the 9-1-1 ring. Biochemical analysis indicates that RAD9 C-tail binds to the hydrophobic pocket on the PCNA-like domain of RAD9, implying that the pocket is involved in multiple protein-protein interactions. The crystal structure of the 9-1-1 ring in complex with a RHINO peptide reveals that RHINO binds to the hydrophobic pocket of RAD9, shedding light on the RAD9-binding motif. Additionally, the study proposes a structural model of the 9-1-1-RHINO quaternary complex. Together, these findings provide functional insights into the intra- and inter-molecular interactions on the front side of RAD9, elucidating the roles of RAD9 C-tail and RHINO in checkpoint activation.
- School of Pharmaceutical Sciences, University of Shizuoka, Shizuoka, Japan.
Organizational Affiliation: