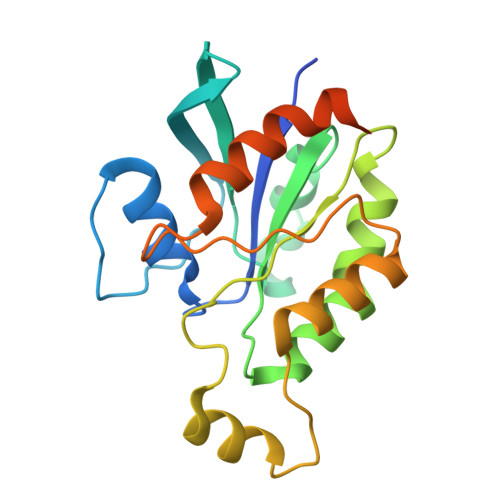
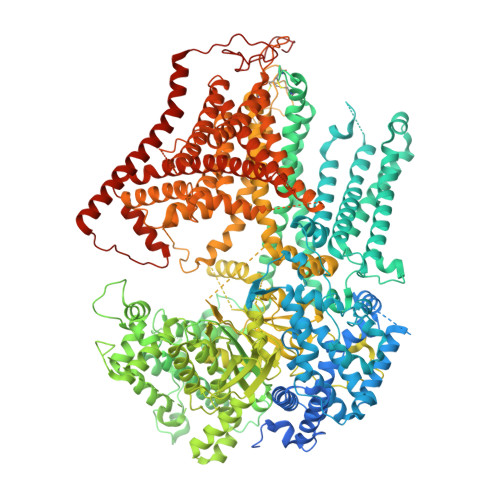
Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex.
Li, J., Liu, H., Li, J., Liu, J., Dai, X., Zhu, A., Xiao, Q., Qian, W., Li, H., Guo, L., Yan, C., Deng, D., Luo, Y., Wang, X.(2025) Nat Commun 16: 2054-2054
- PubMed: 40021629
- DOI: https://doi.org/10.1038/s41467-025-57152-7
- Primary Citation of Related Structures:
8WL6, 8WLA - PubMed Abstract:
β-1,3 Glucan synthase (GS) is essential for fungal cell wall biosynthesis. The GS holoenzyme comprises the glycosyltransferase FKS1 and its regulatory factor Rho1, a small GTPase. However, the mechanism by which Rho1 activates FKS1 in a GTP-dependent manner remains unclear. Here, we present two cryo-EM structures of FKS1, apo and in complex with Rho1. FKS1 adopts a cellulose synthase-like conformation. The interaction between Rho1 and FKS1 is enhanced in the presence of GTPγS. Rho1 is positioned within a pocket between the glycosyltransferase domain of FKS1 (GT domain) and the transmembrane helix spanning TM7-15. Comparison of the two structures reveals extensive conformational changes within FKS1. These alterations suggest that Rho1's GTP/GDP cycling may act as a molecular pump, promoting a dynamic transition between the resting and active states of FKS1. Notably, Rho1 triggers FKS1 conformational changes that may push the growing glucan chain into FKS1's transmembrane channel, thereby facilitating β-1,3-glucan elongation.
- Department of Obstetrics, Key Laboratory of Birth Defects and Related Disease of Women and Children of MOE, State Key Laboratory of Biotherapy, West China Second Hospital, Sichuan University, Chengdu, China.
Organizational Affiliation: