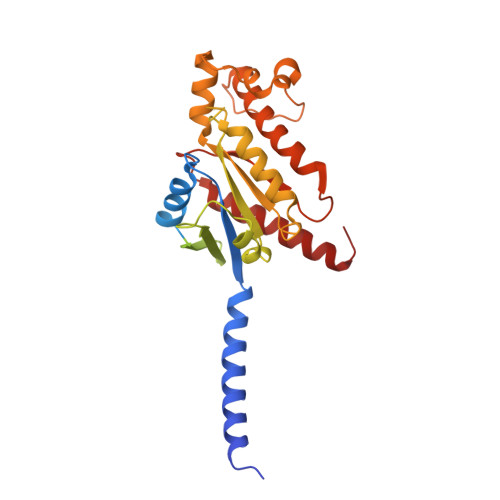
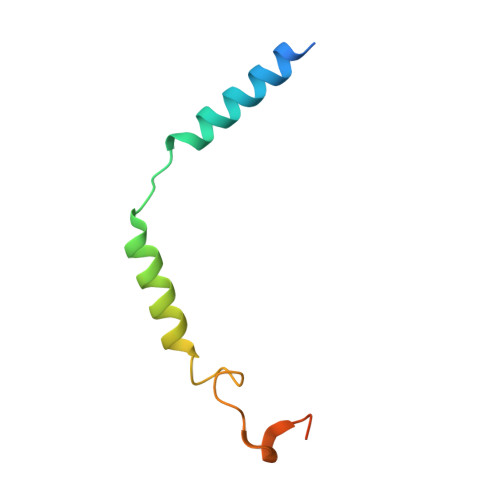
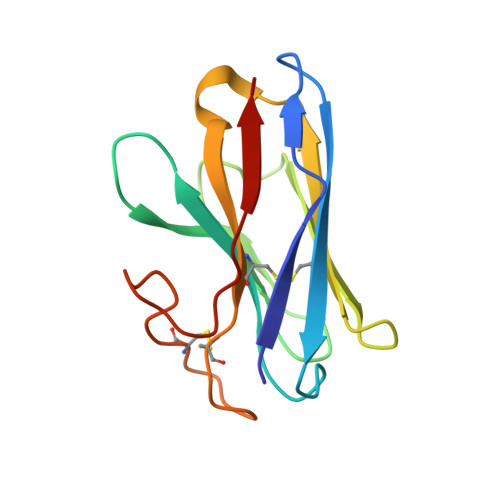
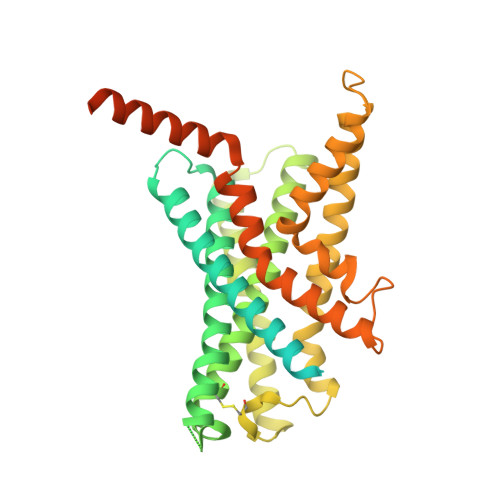
Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cong, Z., Zhao, F., Li, Y., Luo, G., Mai, Y., Chen, X., Chen, Y., Lin, S., Cai, X., Zhou, Q., Yang, D., Wang, M.W.(2024) Cell Discov 10: 18-18
- PubMed: 38346960
- DOI: https://doi.org/10.1038/s41421-024-00649-0
- Primary Citation of Related Structures:
8WA3, 8WG7, 8WG8 - PubMed Abstract:
Class B1 G protein-coupled receptors (GPCRs) are important regulators of many physiological functions such as glucose homeostasis, which is mainly mediated by three peptide hormones, i.e., glucagon-like peptide-1 (GLP-1), glucagon (GCG), and glucose-dependent insulinotropic polypeptide (GIP). They trigger a cascade of signaling events leading to the formation of an active agonist-receptor-G protein complex. However, intracellular signal transducers can also activate the receptor independent of extracellular stimuli, suggesting an intrinsic role of G proteins in this process. Here, we report cryo-electron microscopy structures of the human GLP-1 receptor (GLP-1R), GCG receptor (GCGR), and GIP receptor (GIPR) in complex with G s proteins without the presence of cognate ligands. These ligand-free complexes share a similar intracellular architecture to those bound by endogenous peptides, in which, the G s protein alone directly opens the intracellular binding cavity and rewires the extracellular orthosteric pocket to stabilize the receptor in a state unseen before. While the peptide-binding site is partially occupied by the inward folded transmembrane helix 6 (TM6)-extracellular loop 3 (ECL3) juncture of GIPR or a segment of GCGR ECL2, the extracellular portion of GLP-1R adopts a conformation close to the active state. Our findings offer valuable insights into the distinct activation mechanisms of these three important receptors. It is possible that in the absence of a ligand, the intracellular half of transmembrane domain is mobilized with the help of G s protein, which in turn rearranges the extracellular half to form a transitional conformation, facilitating the entry of the peptide N-terminus.
- Department of Pharmacology, School of Basic Medical Sciences, Fudan University, Shanghai, China.
Organizational Affiliation: