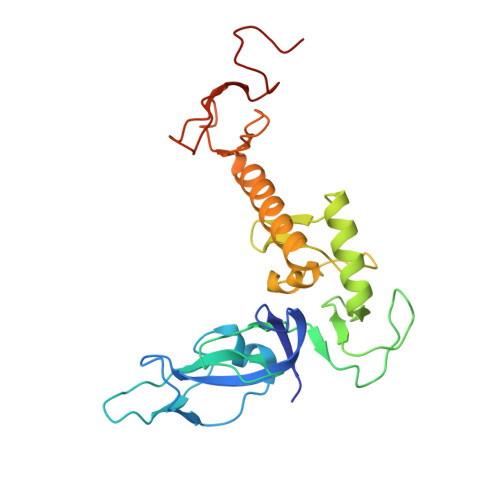
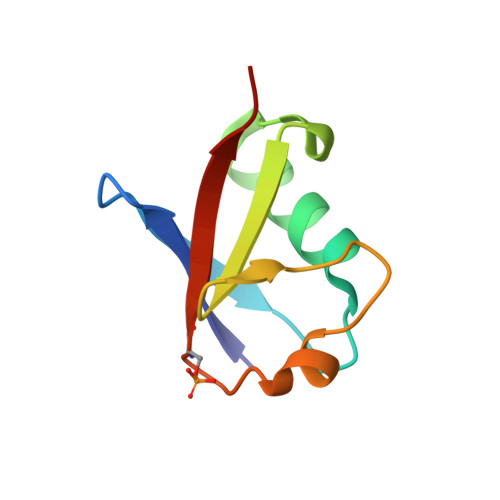
Activation of parkin by a molecular glue.
Sauve, V., Stefan, E., Croteau, N., Goiran, T., Fakih, R., Bansal, N., Hadzipasic, A., Fang, J., Murugan, P., Chen, S., Fon, E.A., Hirst, W.D., Silvian, L.F., Trempe, J.F., Gehring, K.(2024) Nat Commun 15: 7707-7707
- PubMed: 39300082
- DOI: https://doi.org/10.1038/s41467-024-51889-3
- Primary Citation of Related Structures:
8W31 - PubMed Abstract:
Mutations in parkin and PINK1 cause early-onset Parkinson's disease (EOPD). The ubiquitin ligase parkin is recruited to damaged mitochondria and activated by PINK1, a kinase that phosphorylates ubiquitin and the ubiquitin-like domain of parkin. Activated phospho-parkin then ubiquitinates mitochondrial proteins to target the damaged organelle for degradation. Here, we present the mechanism of activation of a new class of small molecule allosteric modulators that enhance parkin activity. The compounds act as molecular glues to enhance the ability of phospho-ubiquitin (pUb) to activate parkin. Ubiquitination assays and isothermal titration calorimetry with the most active compound (BIO-2007817) identify the mechanism of action. We present the crystal structure of a closely related compound (BIO-1975900) bound to a complex of parkin and two pUb molecules. The compound binds next to pUb on RING0 and contacts both proteins. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) experiments confirm that activation occurs through release of the catalytic Rcat domain. In organello and mitophagy assays demonstrate that BIO-2007817 partially rescues the activity of parkin EOPD mutants, R42P and V56E, offering a basis for the design of activators as therapeutics for Parkinson's disease.
- Department of Biochemistry, McGill University, Montreal, QC, Canada.
Organizational Affiliation: