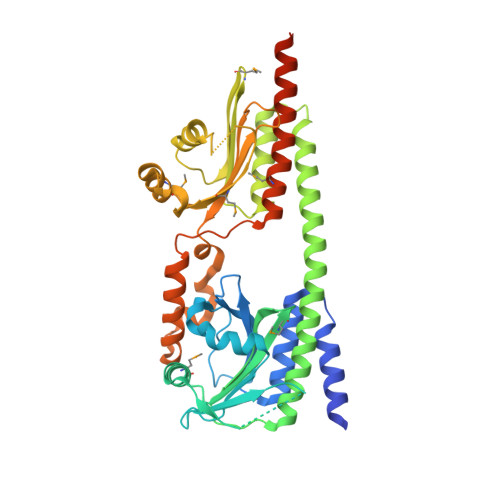
Crystal structure of the photosensory module from a PAS-less cyanobacterial phytochrome as Pr shows a mix of dark-adapted and photoactivated features.
Burgie, E.S., Mickles, A.J., Luo, F., Miller, M.D., Vierstra, R.D.(2024) J Biological Chem 300: 107369-107369
- PubMed: 38750792
- DOI: https://doi.org/10.1016/j.jbc.2024.107369
- Primary Citation of Related Structures:
8W26 - PubMed Abstract:
Phytochromes (Phys) are a diverse collection of photoreceptors that regulate numerous physiological and developmental processes in microorganisms and plants through photointerconversion between red-light-absorbing Pr and far-red light-absorbing Pfr states. Light is detected by an N-terminal photo-sensing module (PSM) sequentially comprised of Period/ARNT/Sim (PAS), cGMP-phosphodiesterase/adenylyl cyclase/FhlA (GAF), and Phy-specific (PHY) domains, with the bilin chromophore covalently-bound within the GAF domain. Phys sense light via the Pr/Pfr ratio measured by the light-induced rotation of the bilin D-pyrrole ring that triggers conformational changes within the PSM, which for microbial Phys reaches into an output region. A key step is a β-stranded to α-helical reconfiguration of a hairpin loop extending from the PHY domain to contact the GAF domain. Besides canonical Phys, cyanobacteria express several variants, including a PAS-less subfamily that harbors just the GAF and PHY domains for light detection. Prior 2D-NMR studies of a model PAS-less Phy from Synechococcus_sp._JA-2-3B'a(2-13) (SyB-Cph1) proposed a unique photoconversion mechanism involving an A-pyrrole ring rotation while magic-angle-spinning NMR probing the chromophore proposed the prototypic D-ring flip. To help solve this conundrum, we determined the crystallographic structure of the GAF-PHY region from SyB-Cph1 as Pr. Surprisingly, this structure differs from canonical Phys by having a Pr ZZZsyn,syn,anti bilin configuration but shifted to the activated position in the binding pocket with consequent folding of the hairpin loop to α-helical, an architecture common for Pfr. Collectively, the PSM of SyB-Cph1 as Pr displayed a mix of dark-adapted and photoactivated features whose co-planar A-C pyrrole rings support a D-ring flip mechanism.
- Department of Biology, Washington University in St Louis, St Louis, Missouri, USA.
Organizational Affiliation: