An adaptive peptide-binding site in ubiquitin receptor hRpn13 revealed by structural studies.
Hassan, B., Chandravanshi, M., Ng, M.Y., Negi, H., Wilson, B.A.P., Walters, K.J.(2025) Nat Commun 16: 5669-5669
- PubMed: 40595513
- DOI: https://doi.org/10.1038/s41467-025-60843-w
- Primary Citation of Related Structures:
8VWO - PubMed Abstract:
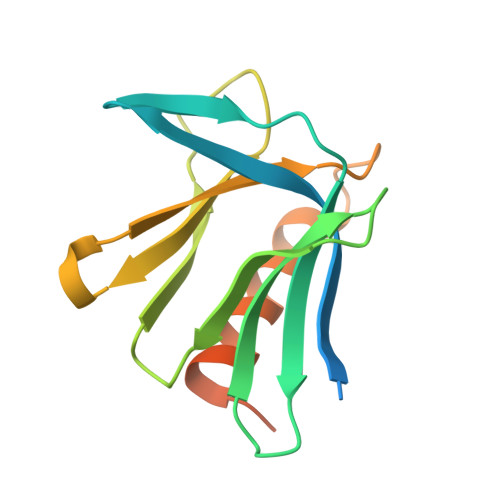
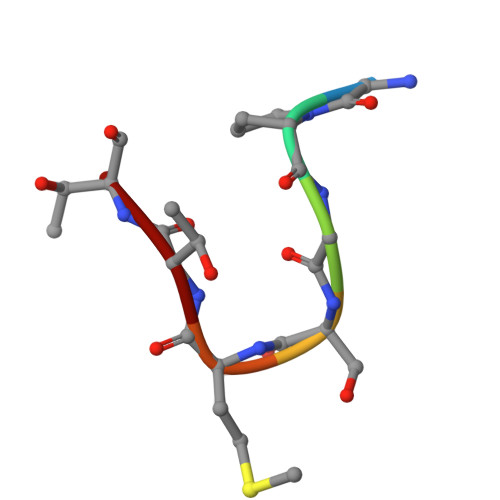
A pleckstrin-like receptor for ubiquitin (Pru) domain in hRpn13 binds ubiquitin and proteasome subunit hRpn2. Here, we report a crystal structure of Pru bound to amino acids at the extreme N-terminus (ENT) of recombinant hRpn13. ENT adopts a U shape with native sequence along one side where M1 is buried in a Pru W108-centered pocket, and non-native sequence along the other with main chain hydrogen bonding to a neighboring Pru of the crystal lattice. These ENT:Pru interactions are stable in molecular dynamics simulations even with inclusion of only one Pru. Our findings suggest that hRpn13 can form bidentate interactions with ubiquitinated substrates by binding to both ubiquitin chains and disordered sequences of substrates. Testing this model by solution nuclear magnetic resonance revealed Pru to bind weakly to various peptides, concurrent binding with ubiquitin, and ENT displacement by hRpn2, the latter required for substrate handoff to the proteasome ATPases.
- Protein Processing Section, Center for Structural Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Organizational Affiliation: