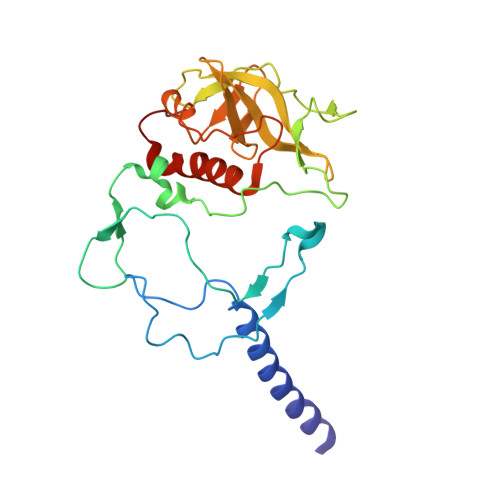
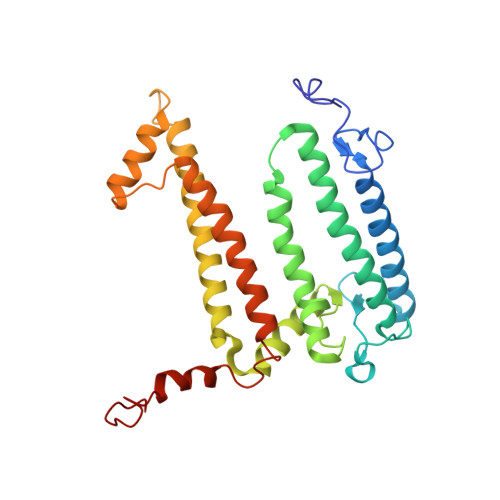
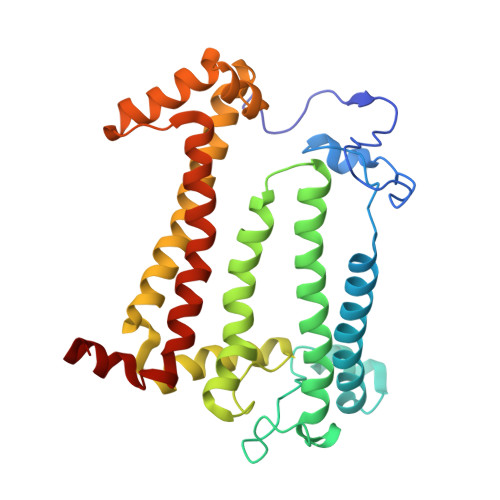
Application of Amber Suppression To Study the Role of Tyr M210 in Electron Transfer in Rhodobacter sphaeroides Photosynthetic Reaction Centers.
Tran, K.N., Faries, K.M., Magdaong, N.C.M., Mathews, I.I., Weaver, J.B., Kirsh, J.M., Holten, D., Kirmaier, C., Boxer, S.G.(2025) J Phys Chem B 129: 3317-3333
- PubMed: 40134359
- DOI: https://doi.org/10.1021/acs.jpcb.5c00082
- Primary Citation of Related Structures:
8VTJ, 8VTK, 8VTL, 8VTM, 8VTN, 8VTO - PubMed Abstract:
The initial light-induced electron transfer (ET) steps in the bacterial photosynthetic reaction center (RC) have been extensively studied and provide a paradigm for connecting structure and function. Although RCs have local pseudo- C 2 symmetry, ET only occurs along the A branch of chromophores. Tyrosine M210 is a key symmetry-breaking residue adjacent to bacteriochlorophyll B A that bridges the primary electron donor P and the bacteriopheophytin acceptor H A . We used amber suppression to incorporate phenylalanine variants with different electron-withdrawing/-donating capabilities at the position M210. X-ray data generally reveal no appreciable structural changes due to the mutations. P* decay and P + H A - formation are multiexponential (∼2 to 9, ∼10 to 60, and ∼100 to 300 ps) and temperature dependent. The 1020 nm transient-absorption band of P + B A - is barely resolved for a few variants at 295 K and for none at 77 K. The results indicate a change from two-step ET for wild-type RCs to the dominance of one-step superexchange ET for the mutants. Resonance Stark spectroscopy reveals that the free energy of P + B A - changes by -57 to +66 meV among the phenylalanine variants. Because P + B A - apparently lies above P* in all phenylalanine variants, the perturbations primarily affect the energy denominator for superexchange mixing. The findings deepen insight into primary ET in the bacterial RC.
- Department of Chemistry, Stanford University, Stanford, California 94305, United States.
Organizational Affiliation: