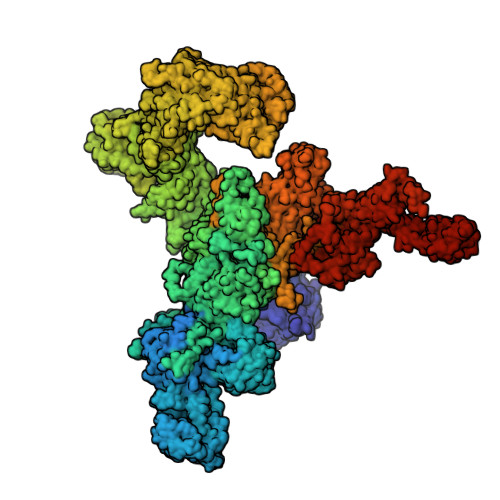
Structural insights into the regulation of RyR1 by S100A1.
Weninger, G., Miotto, M.C., Tchagou, C., Reiken, S., Dridi, H., Brandenburg, S., Riedemann, G.C., Yuan, Q., Liu, Y., Chang, A., Wronska, A., Lehnart, S.E., Marks, A.R.(2024) Proc Natl Acad Sci U S A 121: e2400497121-e2400497121
- PubMed: 38917010
- DOI: https://doi.org/10.1073/pnas.2400497121
- Primary Citation of Related Structures:
8VJJ, 8VJK, 8VK3, 8VK4 - PubMed Abstract:
S100A1, a small homodimeric EF-hand Ca 2+ -binding protein (~21 kDa), plays an important regulatory role in Ca 2+ signaling pathways involved in various biological functions including Ca 2+ cycling and contractile performance in skeletal and cardiac myocytes. One key target of the S100A1 interactome is the ryanodine receptor (RyR), a huge homotetrameric Ca 2+ release channel (~2.3 MDa) of the sarcoplasmic reticulum. Here, we report cryoelectron microscopy structures of S100A1 bound to RyR1, the skeletal muscle isoform, in absence and presence of Ca 2+ . Ca 2+ -free apo-S100A1 binds beneath the bridging solenoid (BSol) and forms contacts with the junctional solenoid and the shell-core linker of RyR1. Upon Ca 2+ -binding, S100A1 undergoes a conformational change resulting in the exposure of the hydrophobic pocket known to serve as a major interaction site of S100A1. Through interactions of the hydrophobic pocket with RyR1, Ca 2+ -bound S100A1 intrudes deeper into the RyR1 structure beneath BSol than the apo-form and induces sideways motions of the C-terminal BSol region toward the adjacent RyR1 protomer resulting in tighter interprotomer contacts. Interestingly, the second hydrophobic pocket of the S100A1-dimer is largely exposed at the hydrophilic surface making it prone to interactions with the local environment, suggesting that S100A1 could be involved in forming larger heterocomplexes of RyRs with other protein partners. Since S100A1 interactions stabilizing BSol are implicated in the regulation of RyR-mediated Ca 2+ release, the characterization of the S100A1 binding site conserved between RyR isoforms may provide the structural basis for the development of therapeutic strategies regarding treatments of RyR-related disorders.
- Department of Physiology and Cellular Biophysics, Center for Molecular Cardiology, Columbia University Vagelos College of Physicians and Surgeons, New York, NY 10032.
Organizational Affiliation: