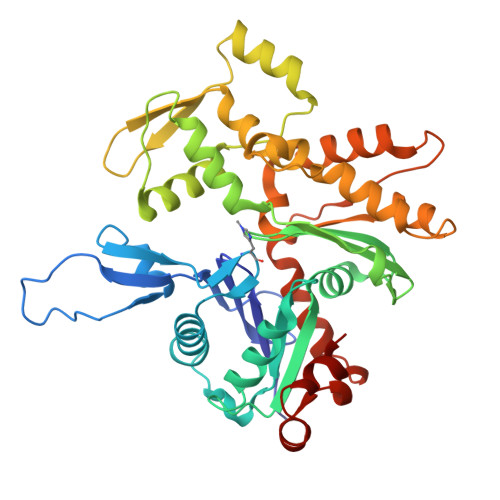
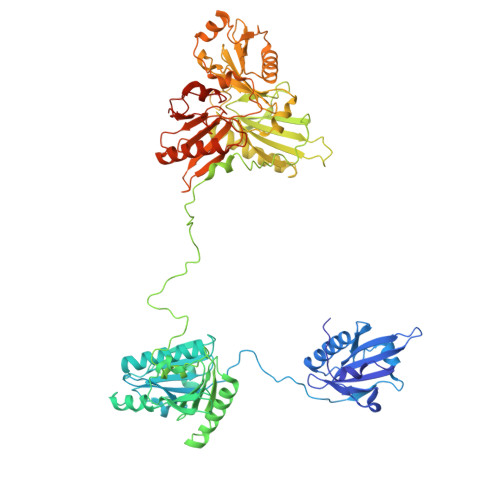
Mechanism of actin filament severing and capping by gelsolin.
Barrie, K.R., Rebowski, G., Dominguez, R.(2025) Nat Struct Mol Biol 32: 237-242
- PubMed: 39448849
- DOI: https://doi.org/10.1038/s41594-024-01412-5
- Primary Citation of Related Structures:
8VIZ, 8VKH - PubMed Abstract:
Gelsolin is the prototypical member of a family of Ca 2+ -activated F-actin severing and capping proteins. Here we report structures of Ca 2+ -bound human gelsolin at the barbed end of F-actin. One structure reveals gelsolin's six domains (G1G6) and interdomain linkers wrapping around F-actin, while another shows domains G1G3-a fragment observed during apoptosis-binding on both sides of F-actin. Conformational changes that trigger severing occur on one side of F-actin with G1G6 and on both sides with G1G3. Gelsolin remains bound after severing, blocking subunit exchange.
- Department of Physiology and Biochemistry and Molecular Biophysics Graduate Group, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Organizational Affiliation: