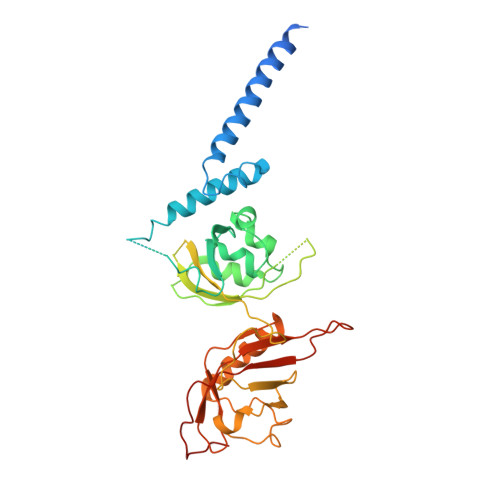
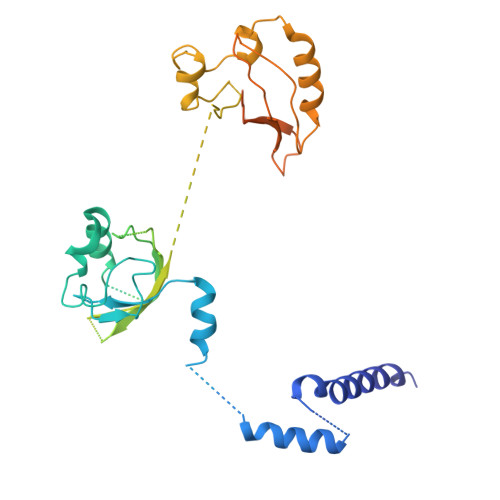
BMAL1-HIF2A heterodimer modulates circadian variations of myocardial injury.
Ruan, W., Li, T., Bang, I.H., Lee, J., Deng, W., Ma, X., Luo, C., Du, F., Yoo, S.H., Kim, B., Li, J., Yuan, X., Figarella, K., An, Y.A., Wang, Y.Y., Liang, Y., DeBerge, M., Zhang, D., Zhou, Z., Wang, Y., Gorham, J.M., Seidman, J.G., Seidman, C.E., Aranki, S.F., Nair, R., Li, L., Narula, J., Zhao, Z., Gorfe, A.A., Muehlschlegel, J.D., Tsai, K.L., Eltzschig, H.K.(2025) Nature 641: 1017-1028
- PubMed: 40269168
- DOI: https://doi.org/10.1038/s41586-025-08898-z
- Primary Citation of Related Structures:
8VHG - PubMed Abstract:
Acute myocardial infarction is a leading cause of morbidity and mortality worldwide 1 . Clinical studies have shown that the severity of cardiac injury after myocardial infarction exhibits a circadian pattern, with larger infarcts and poorer outcomes in patients experiencing morning-onset events 2-7 . However, the molecular mechanisms underlying these diurnal variations remain unclear. Here we show that the core circadian transcription factor BMAL1 7-11 regulates circadian-dependent myocardial injury by forming a transcriptionally active heterodimer with a non-canonical partner-hypoxia-inducible factor 2 alpha (HIF2A) 12-16 -in a diurnal manner. To substantiate this finding, we determined the cryo-EM structure of the BMAL1-HIF2A-DNA complex, revealing structural rearrangements within BMAL1 that enable cross-talk between circadian rhythms and hypoxia signalling. BMAL1 modulates the circadian hypoxic response by enhancing the transcriptional activity of HIF2A and stabilizing the HIF2A protein. We further identified amphiregulin (AREG) 16,17 as a rhythmic target of the BMAL1-HIF2A complex, critical for regulating daytime variations of myocardial injury. Pharmacologically targeting the BMAL1-HIF2A-AREG pathway provides cardioprotection, with maximum efficacy when aligned with the pathway's circadian phase. These findings identify a mechanism governing circadian variations of myocardial injury and highlight the therapeutic potential of clock-based pharmacological interventions for treating ischaemic heart disease.
- Department of Anesthesiology, Critical Care and Pain Medicine, The University of Texas Health Science Center at Houston, McGovern Medical School, Houston, TX, USA. Wei.Ruan@uth.tmc.edu.
Organizational Affiliation: