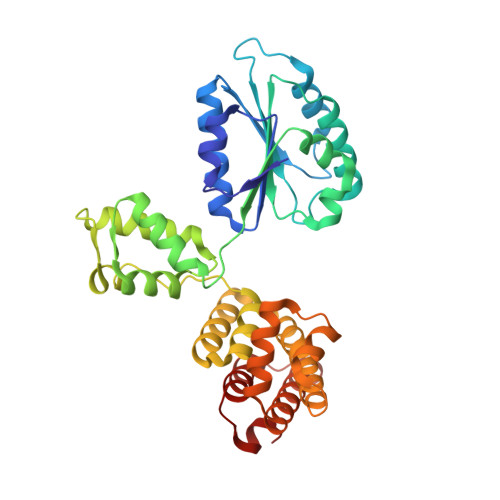
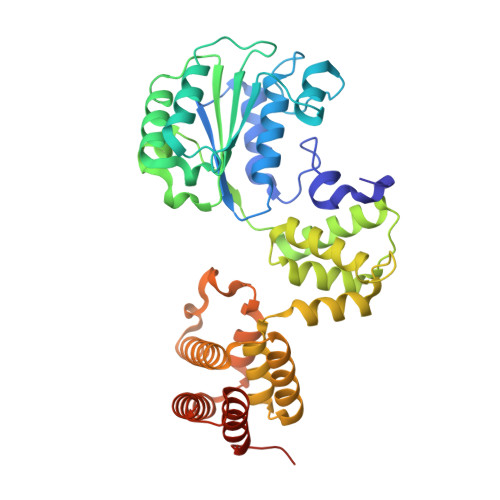
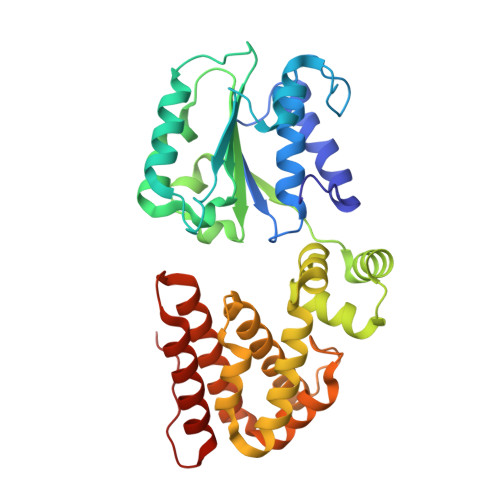
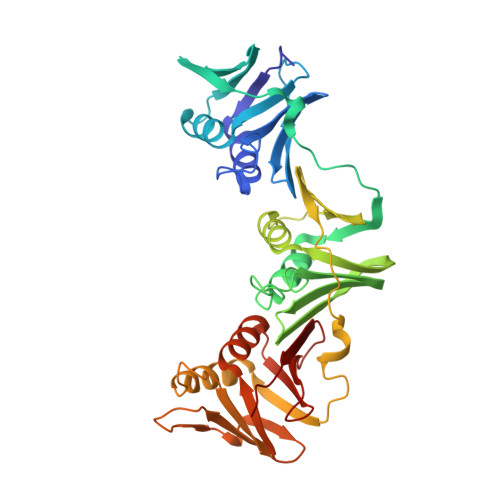
Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication.
Landeck, J.T., Pajak, J., Norman, E.K., Sedivy, E.L., Kelch, B.A.(2024) J Biological Chem 300: 107166-107166
- PubMed: 38490435
- DOI: https://doi.org/10.1016/j.jbc.2024.107166
- Primary Citation of Related Structures:
8VAL, 8VAM, 8VAN, 8VAP, 8VAQ, 8VAR, 8VAS, 8VAT - PubMed Abstract:
Clamp loaders are pentameric ATPases that place circular sliding clamps onto DNA, where they function in DNA replication and genome integrity. The central activity of a clamp loader is the opening of the ring-shaped sliding clamp and the subsequent binding to primer-template (p/t)-junctions. The general architecture of clamp loaders is conserved across all life, suggesting that their mechanism is retained. Recent structural studies of the eukaryotic clamp loader replication factor C (RFC) revealed that it functions using a crab-claw mechanism, where clamp opening is coupled to a massive conformational change in the loader. Here we investigate the clamp loading mechanism of the Escherichia coli clamp loader at high resolution using cryo-electron microscopy. We find that the E. coli clamp loader opens the clamp using a crab-claw motion at a single pivot point, whereas the eukaryotic RFC loader uses motions distributed across the complex. Furthermore, we find clamp opening occurs in multiple steps, starting with a partly open state with a spiral conformation, and proceeding to a wide open clamp in a surprising planar geometry. Finally, our structures in the presence of p/t-junctions illustrate how the clamp closes around p/t-junctions and how the clamp loader initiates release from the loaded clamp. Our results reveal mechanistic distinctions in a macromolecular machine that is conserved across all domains of life.
- Department of Biochemistry and Molecular Biotechnology, University of Massachusetts Chan Medical School, Worcester, Massachusetts, USA.
Organizational Affiliation: