Cryo-EM structure revealed a novel F-actin binding motif in a Legionella pneumophila lysine fatty-acyltransferase
Zeng, W.W., Komaniecki, G., Liu, J., Lin, H., Mao, Y.(2025) Elife
Experimental Data Snapshot
Starting Models: in silico
View more details
wwPDB Validation 3D Report Full Report
(2025) Elife
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin, alpha skeletal muscle | 370 | Bos taurus | Mutation(s): 0 EC: 3.6.4 | 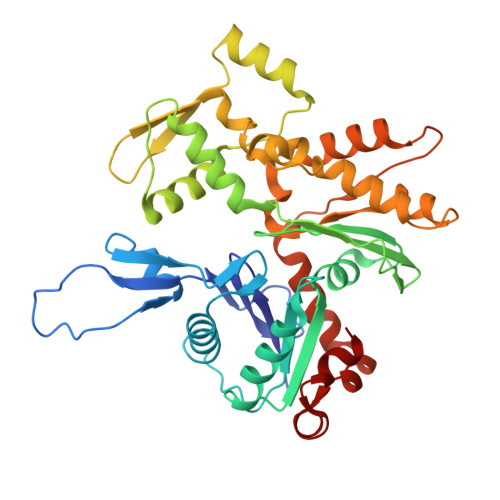 | |
UniProt | |||||
Find proteins for P68138 (Bos taurus) Explore P68138 Go to UniProtKB: P68138 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68138 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| F-actin-binding Coiled-Coil domain of LFAT1 | 69 | Legionella pneumophila | Mutation(s): 0 Gene Names: lpg1387 | 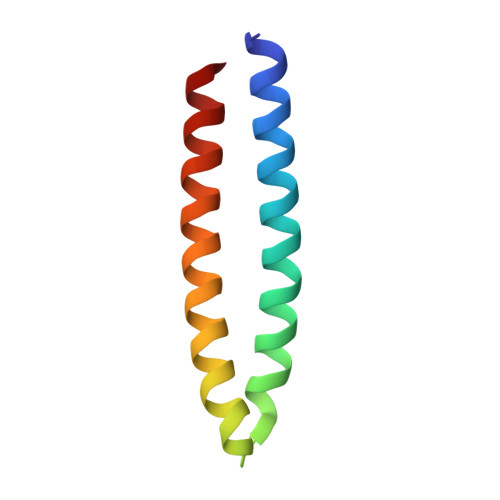 | |
UniProt | |||||
Find proteins for Q5ZVQ3 (Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)) Explore Q5ZVQ3 Go to UniProtKB: Q5ZVQ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ZVQ3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | AA [auth D] CA [auth E] EA [auth F] GA [auth G] IA [auth H] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | BA [auth D] DA [auth E] FA [auth F] HA [auth G] JA [auth H] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3.0 |
| MODEL REFINEMENT | PHENIX | 9.0 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01-GM135379-01 |