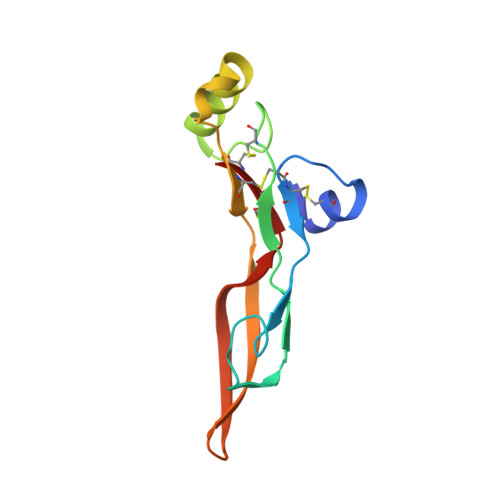
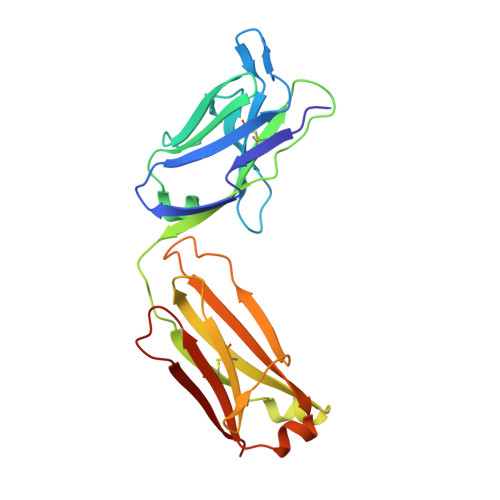
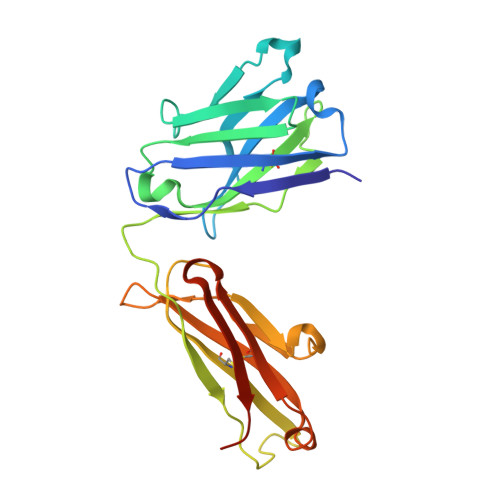
Isoform-selective TGF-beta 3 inhibition for systemic sclerosis.
Sun, T., Vander Heiden, J.A., Gao, X., Yin, J., Uttarwar, S., Liang, W.C., Jia, G., Yadav, R., Huang, Z., Mitra, M., Halpern, W., Bender, H.S., Brightbill, H.D., Wu, Y., Lupardus, P., Ramalingam, T., Arron, J.R.(2024) Med 5: 132-147.e7
- PubMed: 38272035
- DOI: https://doi.org/10.1016/j.medj.2023.12.011
- Primary Citation of Related Structures:
8V52 - PubMed Abstract:
Transforming growth factor β (TGF-β) is implicated as a key mediator of pathological fibrosis, but its pleiotropic activity in a range of homeostatic functions presents challenges to its safe and effective therapeutic targeting. There are three isoforms of TGF-β, TGF-β1, TGF-β2, and TGF-β3, which bind to a common receptor complex composed of TGF-βR1 and TGF-βR2 to induce similar intracellular signals in vitro. We have recently shown that the cellular expression patterns and activation thresholds of TGF-β2 and TGF-β3 are distinct from those of TGF-β1 and that selective short-term TGF-β2 and TGF-β3 inhibition can attenuate fibrosis in vivo without promoting excessive inflammation. Isoform-selective inhibition of TGF-β may therefore provide a therapeutic opportunity for patients with chronic fibrotic disorders. Transcriptomic profiling of skin biopsies from patients with systemic sclerosis (SSc) from multiple clinical trials was performed to evaluate the role of TGF-β3 in this disease. Antibody humanization, biochemical characterization, crystallization, and pre-clinical experiments were performed to further characterize an anti-TGF-β3 antibody. In the skin of patients with SSc, TGF-β3 expression is uniquely correlated with biomarkers of TGF-β signaling and disease severity. Crystallographic studies establish a structural basis for selective TGF-β3 inhibition with a potent and selective monoclonal antibody that attenuates fibrosis effectively in vivo at clinically translatable exposures. Toxicology studies suggest that, as opposed to pan-TGF-β inhibitors, this anti-TGF-β3 antibody has a favorable safety profile for chronic administration. We establish a rationale for targeting TGF-β3 in SSc with a favorable therapeutic index. This study was funded by Genentech, Inc.
- Department of Immunology, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA. Electronic address: sun.tianhe@gene.com.
Organizational Affiliation: