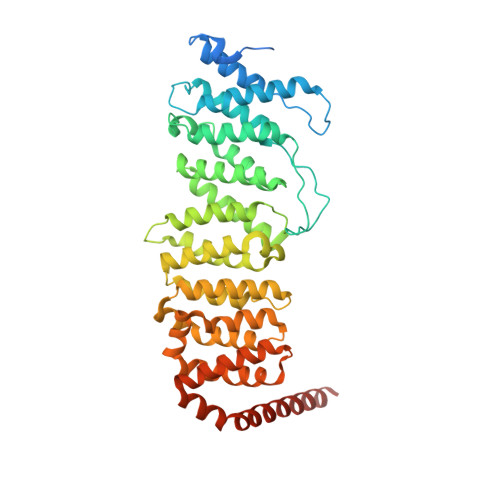
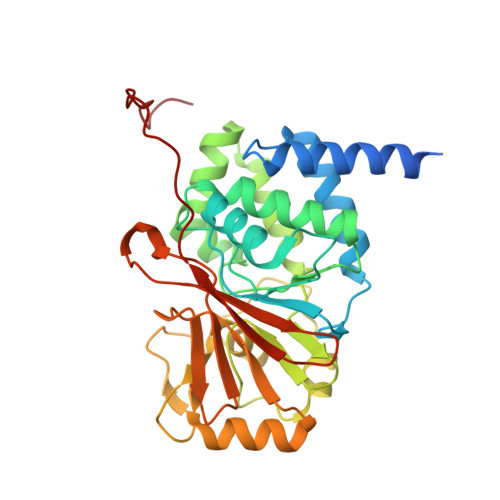
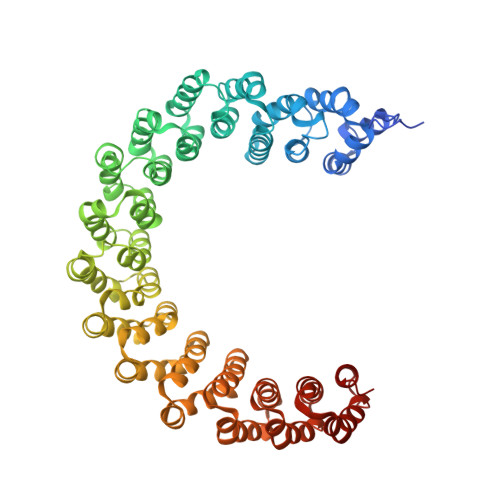Structural characterization of methylation-independent PP2A assembly guides alphafold2Multimer prediction of family-wide PP2A complexes.
Wachter, F., Nowak, R.P., Ficarro, S., Marto, J., Fischer, E.S.(2024) J Biological Chem 300: 107268-107268
- PubMed: 38582449
- DOI: https://doi.org/10.1016/j.jbc.2024.107268
- Primary Citation of Related Structures:
8UWB - PubMed Abstract:
Dysregulation of phosphorylation-dependent signaling is a hallmark of tumorigenesis. Protein phosphatase 2 (PP2A) is an essential regulator of cell growth. One scaffold subunit (A) binds to a catalytic subunit (C) to form a core AC heterodimer, which together with one of many regulatory (B) subunits forms the active trimeric enzyme. The combinatorial number of distinct PP2A complexes is large, which results in diverse substrate specificity and subcellular localization. The detailed mechanism of PP2A assembly and regulation remains elusive and reports about an important role of methylation of the carboxy terminus of PP2A C are conflicting. A better understanding of the molecular underpinnings of PP2A assembly and regulation is critical to dissecting PP2A function in physiology and disease. Here, we combined biochemical reconstitution, mass spectrometry, X-ray crystallography, and functional assays to characterize the assembly of trimeric PP2A. In vitro studies demonstrated that methylation of the carboxy-terminus of PP2A C was dispensable for PP2A assembly in vitro. To corroborate these findings, we determined the X-ray crystal structure of the unmethylated PP2A Aα-B56ε-Cα trimer complex to 3.1 Å resolution. The experimental structure superimposed well with an Alphafold2Multimer prediction of the PP2A trimer. We then predicted models of all canonical PP2A complexes providing a framework for structural analysis of PP2A. In conclusion, methylation was dispensable for trimeric PP2A assembly and integrative structural biology studies of PP2A offered predictive models for all canonical PP2A complexes.
- Department of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA; Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts, USA.
Organizational Affiliation: