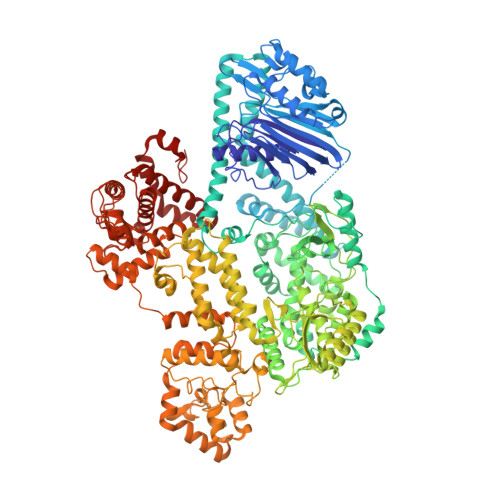
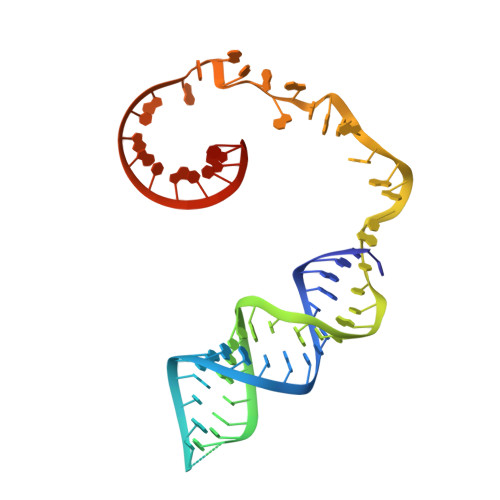
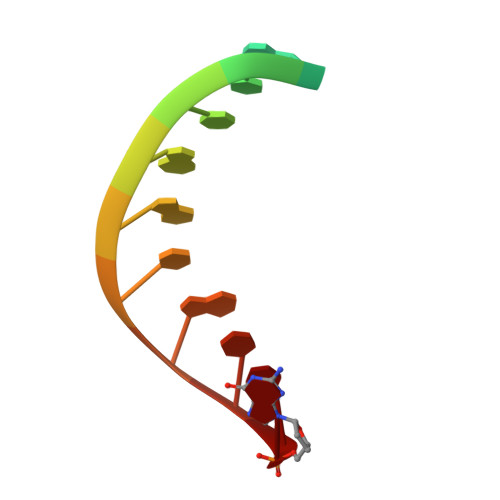
Template and target-site recognition by human LINE-1 in retrotransposition.
Thawani, A., Ariza, A.J.F., Nogales, E., Collins, K.(2024) Nature 626: 186-193
- PubMed: 38096901
- DOI: https://doi.org/10.1038/s41586-023-06933-5
- Primary Citation of Related Structures:
8UW3 - PubMed Abstract:
The long interspersed element-1 (LINE-1, hereafter L1) retrotransposon has generated nearly one-third of the human genome and serves as an active source of genetic diversity and human disease 1 . L1 spreads through a mechanism termed target-primed reverse transcription, in which the encoded enzyme (ORF2p) nicks the target DNA to prime reverse transcription of its own or non-self RNAs 2 . Here we purified full-length L1 ORF2p and biochemically reconstituted robust target-primed reverse transcription with template RNA and target-site DNA. We report cryo-electron microscopy structures of the complete human L1 ORF2p bound to structured template RNAs and initiating cDNA synthesis. The template polyadenosine tract is recognized in a sequence-specific manner by five distinct domains. Among them, an RNA-binding domain bends the template backbone to allow engagement of an RNA hairpin stem with the L1 ORF2p C-terminal segment. Moreover, structure and biochemical reconstitutions demonstrate an unexpected target-site requirement: L1 ORF2p relies on upstream single-stranded DNA to position the adjacent duplex in the endonuclease active site for nicking of the longer DNA strand, with a single nick generating a staggered DNA break. Our research provides insights into the mechanism of ongoing transposition in the human genome and informs the engineering of retrotransposon proteins for gene therapy.
- California Institute for Quantitative Biosciences (QB3), Berkeley, CA, USA. athawani@berkeley.edu.
Organizational Affiliation: