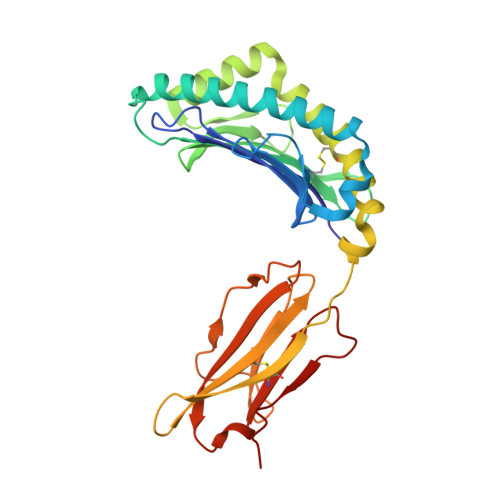
Structure of the murine CD94-NKG2A receptor in complex with Qa-1 b presenting an MHC-I leader peptide.
MacLachlan, B.J., Sullivan, L.C., Brooks, A.G., Rossjohn, J., Vivian, J.P.(2024) FEBS J 291: 1530-1544
- PubMed: 38158698
- DOI: https://doi.org/10.1111/febs.17050
- Primary Citation of Related Structures:
8UMO - PubMed Abstract:
The heterodimeric natural killer cells antigen CD94 (CD94)-NKG2-A/NKG2-B type II integral membrane protein (NKG2A) receptor family expressed on human and mouse natural killer (NK) cells monitors global major histocompatibility complex (MHC) class I cell surface expression levels through binding to MHC class Ia-derived leader sequence peptides presented by HLA class I histocompatibility antigen, alpha chain E (HLA-E; in humans) or H-2 class I histocompatibility antigen, D-37 (Qa-1 b ; in mice). Although the molecular basis underpinning human CD94-NKG2A recognition of HLA-E is known, the equivalent interaction in the murine setting is not. By determining the high-resolution crystal structure of murine CD94-NKG2A in complex with Qa-1 b presenting the Qa-1 determinant modifier peptide (QDM), we resolved the mode of binding. Compared to the human homologue, the murine CD94-NKG2A-Qa-1 b -QDM displayed alterations in the distribution of interactions across CD94 and NKG2A subunits that coincide with differences in electrostatic complementarity of the ternary complex and the lack of cross-species reactivity. Nevertheless, we show that Qa-1b could be modified through W65R + N73I mutations to mimic HLA-E, facilitating binding with both human and murine CD94-NKG2A. These data underscore human and murine CD94-NKG2A cross-species heterogeneity and provide a foundation for humanising Qa-1b in immune system models.
- Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Australia.
Organizational Affiliation: