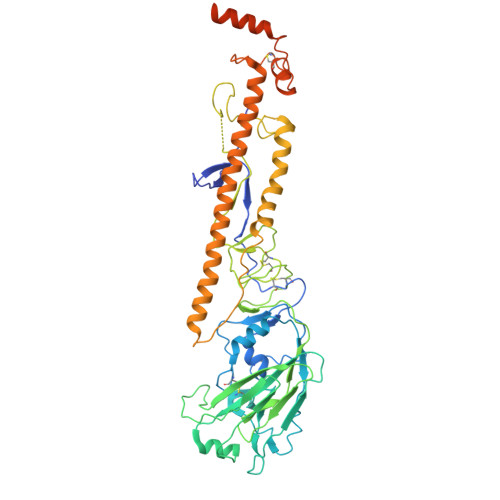
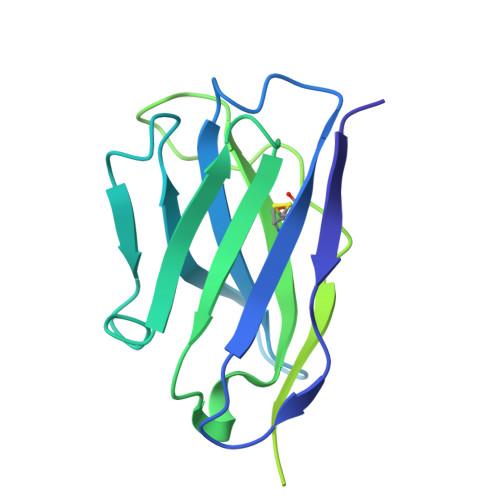
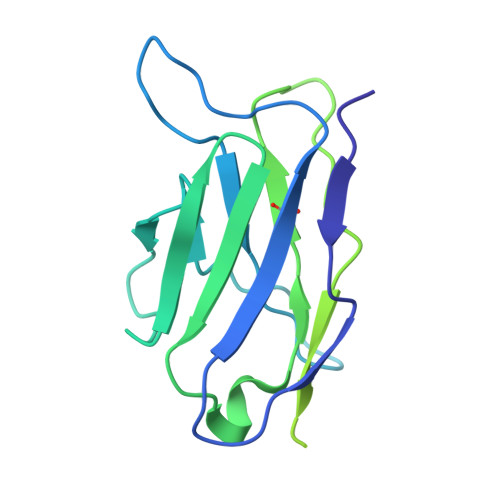
Human naive B cells recognize prepandemic influenza virus hemagglutinins.
Feldman, J., Ramos, A.S.F., Vu, M., Maurer, D.P., Rosado, V.C., Lingwood, D., Bajic, G., Schmidt, A.G.(2025) Sci Immunol 10: eado9572-eado9572
- PubMed: 39854479
- DOI: https://doi.org/10.1126/sciimmunol.ado9572
- Primary Citation of Related Structures:
8UME - PubMed Abstract:
Understanding the naïve B cell repertoire and its specificity for potential zoonotic threats, such as the highly pathogenic avian influenza (HPAI) H5Nx viruses, may allow prediction of infection- or vaccine-specific responses. However, this naïve repertoire and the possibility to respond to emerging, prepandemic viruses are largely undetermined. Here, we profiled naïve B cell reactivity against a prototypical HPAI H5 hemagglutinin (HA), the major target of antibody responses. We found that the frequency of H5-specific human naïve B cells targeting the HA "head" domain was increased relative to cross-reactive B cells to a circulating seasonal H1N1 strain. We classified the isolated monoclonal antibodies (mAbs) by the HA epitopes engaged and found that selected mAbs neutralized H5N1 at germline. We determined a cryo-electron microscopic structure of one mAb in complex with H5 HA to define its epitope. Our study defines the naïve human B cell repertoire recognizing a potentially zoonotic HPAI.
- Ragon Institute of Mass General, MIT, and Harvard, Cambridge, MA 02139, USA.
Organizational Affiliation: