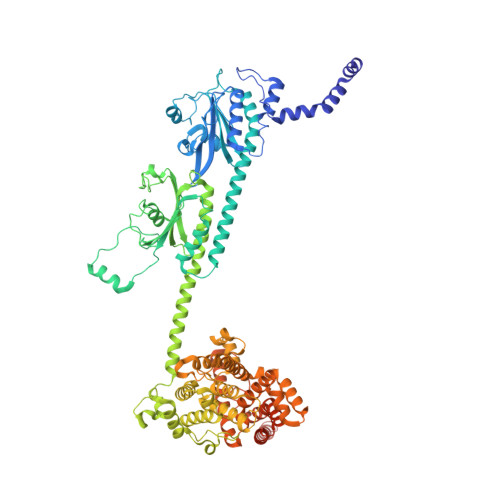
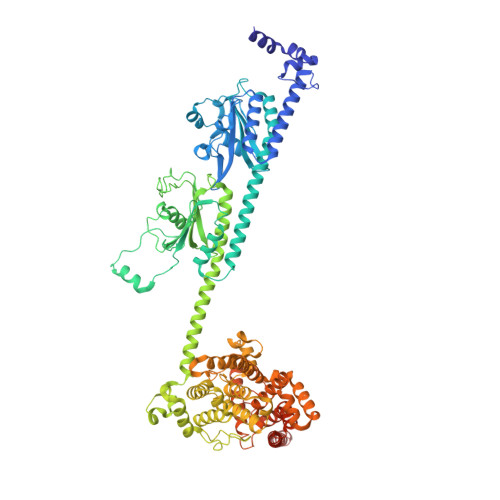
Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
Aplin, C., Cerione, R.A.(2023) J Biological Chem 300: 105608-105608
- PubMed: 38159849
- DOI: https://doi.org/10.1016/j.jbc.2023.105608
- Primary Citation of Related Structures:
8UFI, 8UGB, 8UGS, 8ULG - PubMed Abstract:
Phototransduction in retinal rods occurs when the G protein-coupled photoreceptor rhodopsin triggers the activation of phosphodiesterase 6 (PDE6) by GTP-bound alpha subunits of the G protein transducin (Gα T ). Recently, we presented a cryo-EM structure for a complex between two GTP-bound recombinant Gα T subunits and native PDE6, that included a bivalent antibody bound to the C-terminal ends of Gα T and the inhibitor vardenafil occupying the active sites on the PDEα and PDEβ subunits. We proposed Gα T -activated PDE6 by inducing a striking reorientation of the PDEγ subunits away from the catalytic sites. However, questions remained including whether in the absence of the antibody Gα T binds to PDE6 in a similar manner as observed when the antibody is present, does Gα T activate PDE6 by enabling the substrate cGMP to access the catalytic sites, and how does the lipid membrane enhance PDE6 activation? Here, we demonstrate that 2:1 Gα T -PDE6 complexes form with either recombinant or retinal Gα T in the absence of the Gα T antibody. We show that Gα T binding is not necessary for cGMP nor competitive inhibitors to access the active sites; instead, occupancy of the substrate binding sites enables Gα T to bind and reposition the PDE6γ subunits to promote catalytic activity. Moreover, we demonstrate by reconstituting Gα T -stimulated PDE6 activity in lipid bilayer nanodiscs that the membrane-induced enhancement results from an increase in the apparent binding affinity of Gα T for PDE6. These findings provide new insights into how the retinal G protein stimulates rapid catalytic turnover by PDE6 required for dim light vision.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York, USA.
Organizational Affiliation: