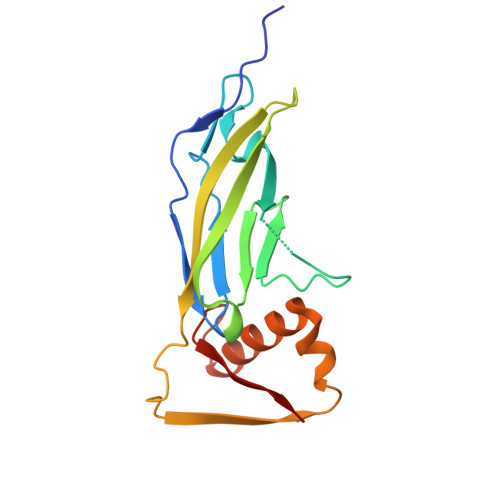
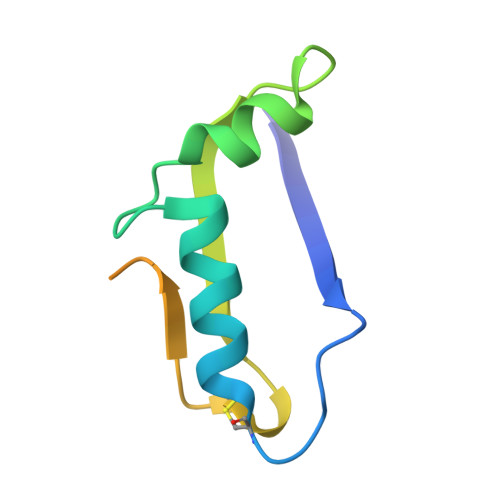
Molecular basis of proteolytic cleavage regulation by the extracellular matrix receptor dystroglycan.
Anderson, M.J.M., Hayward, A.N., Smiley, A.T., Shi, K., Pawlak, M.R., Aird, E.J., Grant, E., Greenberg, L., Aihara, H., Evans 3rd, R.L., Ulens, C., Gordon, W.R.(2024) Structure 32: 1984-1996.e5
- PubMed: 39305901
- DOI: https://doi.org/10.1016/j.str.2024.08.019
- Primary Citation of Related Structures:
8UF4 - PubMed Abstract:
The dystrophin-glycoprotein-complex (DGC), anchored by the transmembrane protein dystroglycan, functions to mechanically link the extracellular matrix and actin cytoskeleton. Breaking this connection is associated with diseases such as muscular dystrophy, yet cleavage of dystroglycan by matrix-metalloproteinases (MMPs) remains an understudied mechanism to disrupt the DGC. We determined the crystal structure of the membrane-adjacent domain (amino acids 491-722) of E. coli expressed human dystroglycan to understand MMP cleavage regulation. The structural model includes tandem immunoglobulin-like (IGL) and sperm/enterokinase/agrin-like (SEAL) domains, which support proteolysis in diverse receptors to facilitate mechanotransduction, membrane protection, and viral entry. The structure reveals a C-terminal extension that buries the MMP site by packing into a hydrophobic pocket, a unique mechanism of MMP cleavage regulation. We further demonstrate structure-guided and disease-associated mutations disrupt proteolytic regulation using a cell-surface proteolysis assay. Thus disrupted proteolysis is a potentially relevant mechanism for "breaking" the DGC link to contribute to disease pathogenesis.
- Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota-Twin Cities, Minneapolis, MN 55455, USA.
Organizational Affiliation: