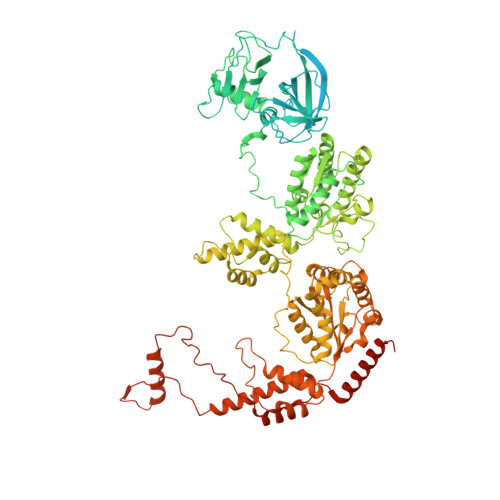
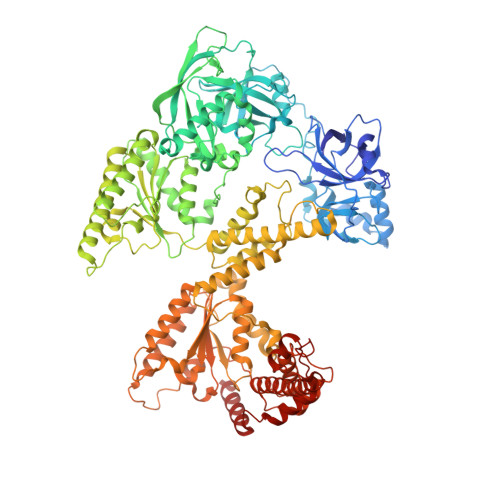
The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.
Ali, B.A., Judy, R.M., Chowdhury, S., Jacobsen, N.K., Castanzo, D.T., Carr, K.L., Richardson, C.D., Lander, G.C., Martin, A., Gardner, B.M.(2023) J Biological Chem 300: 105504-105504
- PubMed: 38036174
- DOI: https://doi.org/10.1016/j.jbc.2023.105504
- Primary Citation of Related Structures:
8U0V, 8U0X - PubMed Abstract:
The heterohexameric ATPases associated with diverse cellular activities (AAA)-ATPase Pex1/Pex6 is essential for the formation and maintenance of peroxisomes. Pex1/Pex6, similar to other AAA-ATPases, uses the energy from ATP hydrolysis to mechanically thread substrate proteins through its central pore, thereby unfolding them. In related AAA-ATPase motors, substrates are recruited through binding to the motor's N-terminal domains or N terminally bound cofactors. Here, we use structural and biochemical techniques to characterize the function of the N1 domain in Pex6 from budding yeast, Saccharomyces cerevisiae. We found that although Pex1/ΔN1-Pex6 is an active ATPase in vitro, it does not support Pex1/Pex6 function at the peroxisome in vivo. An X-ray crystal structure of the isolated Pex6 N1 domain shows that the Pex6 N1 domain shares the same fold as the N-terminal domains of PEX1, CDC48, and NSF, despite poor sequence conservation. Integrating this structure with a cryo-EM reconstruction of Pex1/Pex6, AlphaFold2 predictions, and biochemical assays shows that Pex6 N1 mediates binding to both the peroxisomal membrane tether Pex15 and an extended loop from the D2 ATPase domain of Pex1 that influences Pex1/Pex6 heterohexamer stability. Given the direct interactions with both Pex15 and the D2 ATPase domains, the Pex6 N1 domain is poised to coordinate binding of cofactors and substrates with Pex1/Pex6 ATPase activity.
- Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Barbara, California, USA.
Organizational Affiliation: