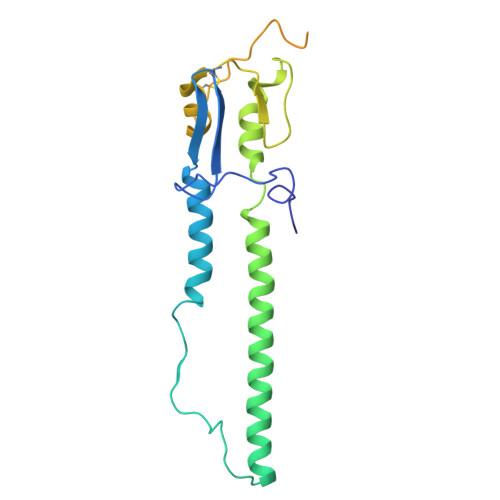
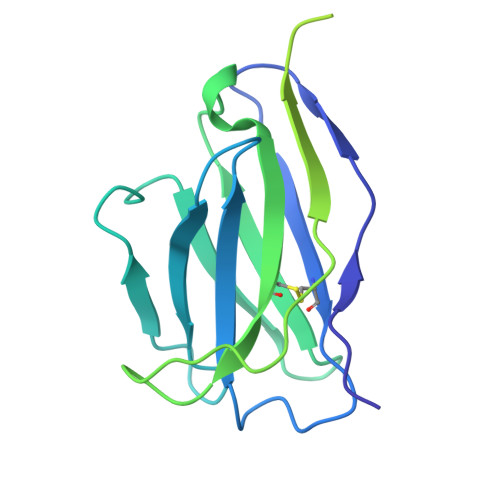
The N terminus of H3-influenza hemagglutinin as a site-of-vulnerability to neutralizing antibody
Rawi, R., Morano, N.C., Cheung, C.S.F., Du, H., Gorman, J., Prabhakaran, M., Becker, J.E., Bylund, T., Charaf, S., Chen, X., Lee, M., Harris, D.R., Olia, A.S., Ou, L., Wang, L., Wang, S., Zhang, B., Kanekiyo, M., McDermott, A.B., Zhou, T., Shapiro, L., Kwong, P.D.(2025) Structure