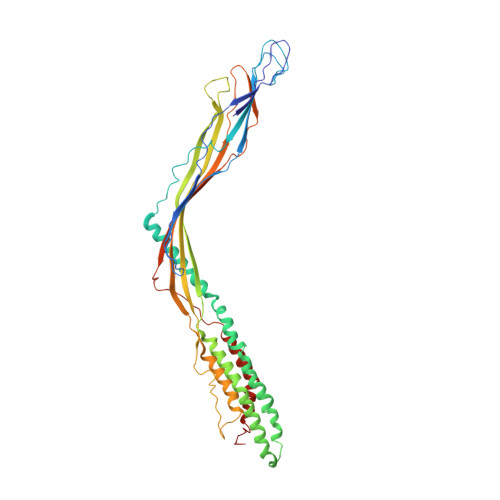
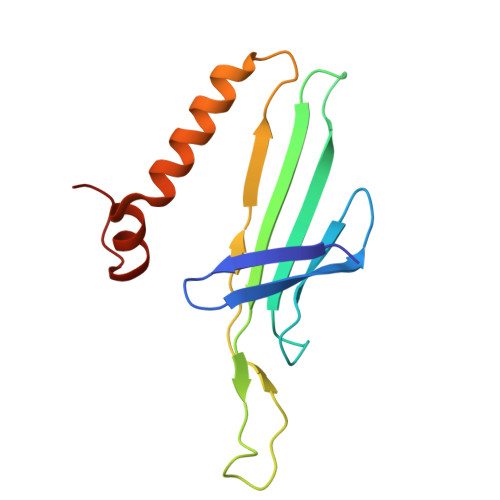
Removal of Pseudomonas type IV pili by a small RNA virus.
Thongchol, J., Yu, Z., Harb, L., Lin, Y., Koch, M., Theodore, M., Narsaria, U., Shaevitz, J., Gitai, Z., Wu, Y., Zhang, J., Zeng, L.(2024) Science 384: eadl0635-eadl0635
- PubMed: 38574145
- DOI: https://doi.org/10.1126/science.adl0635
- Primary Citation of Related Structures:
8TUM, 8TUW, 8TUX - PubMed Abstract:
The retractile type IV pilus (T4P) is important for virulence of the opportunistic human pathogen Pseudomonas aeruginosa . The single-stranded RNA (ssRNA) phage PP7 binds to T4P and is brought to the cell surface through pilus retraction. Using fluorescence microscopy, we discovered that PP7 detaches T4P, which impairs cell motility and restricts the pathogen's virulence. Using cryo-electron microscopy, mutagenesis, optical trapping, and Langevin dynamics simulation, we resolved the structure of PP7, T4P, and the PP7/T4P complex and showed that T4P detachment is driven by the affinity between the phage maturation protein and its bound pilin, plus the pilus retraction force and speed, and pilus bending. Pilus detachment may be widespread among other ssRNA phages and their retractile pilus systems and offers new prospects for antibacterial prophylaxis and therapeutics.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, USA.
Organizational Affiliation: