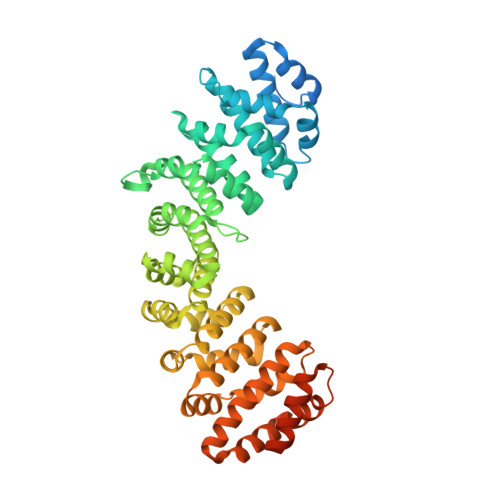
A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Cross, E.M., Akbari, N., Ghassabian, H., Hoad, M., Pavan, S., Ariawan, D., Donnelly, C.M., Lavezzo, E., Petersen, G.F., Forwood, J.K., Alvisi, G.(2024) Protein Sci 33: e4876-e4876
- PubMed: 38108201
- DOI: https://doi.org/10.1002/pro.4876
- Primary Citation of Related Structures:
8Q8K, 8SUD, 8TUQ, 8TUR, 8TUS, 8TUT, 8TUU, 8TUV - PubMed Abstract:
Nucleocytoplasmic transport regulates the passage of proteins between the nucleus and cytoplasm. In the best characterized pathway, importin (IMP) α bridges cargoes bearing basic, classical nuclear localization signals (cNLSs) to IMPβ1, which mediates transport through the nuclear pore complex. IMPα recognizes three types of cNLSs via two binding sites: the major binding site accommodates monopartite cNLSs, the minor binding site recognizes atypical cNLSs, while bipartite cNLSs simultaneously interact with both major and minor sites. Despite the growing knowledge regarding IMPα-cNLS interactions, our understanding of the evolution of cNLSs is limited. We combined bioinformatic, biochemical, functional, and structural approaches to study this phenomenon, using polyomaviruses (PyVs) large tumor antigens (LTAs) as a model. We characterized functional cNLSs from all human (H)PyV LTAs, located between the LXCXE motif and origin binding domain. Surprisingly, the prototypical SV40 monopartite NLS is not well conserved; HPyV LTA NLSs are extremely heterogenous in terms of structural organization, IMPα isoform binding, and nuclear targeting abilities, thus influencing the nuclear accumulation properties of full-length proteins. While several LTAs possess bipartite cNLSs, merkel cell PyV contains a hybrid bipartite cNLS whose upstream stretch of basic amino acids can function as an atypical cNLS, specifically binding to the IMPα minor site upon deletion of the downstream amino acids after viral integration in the host genome. Therefore, duplication of a monopartite cNLS and subsequent accumulation of point mutations, optimizing interaction with distinct IMPα binding sites, led to the evolution of bipartite and atypical NLSs binding at the minor site.
- School of Dentistry and Medical Sciences, Charles Sturt University, Wagga Wagga, Australia.
Organizational Affiliation: