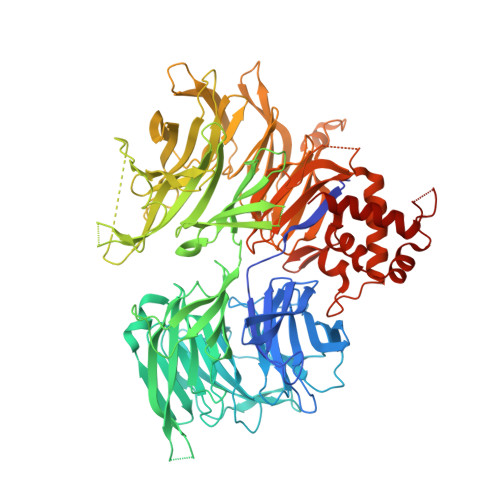
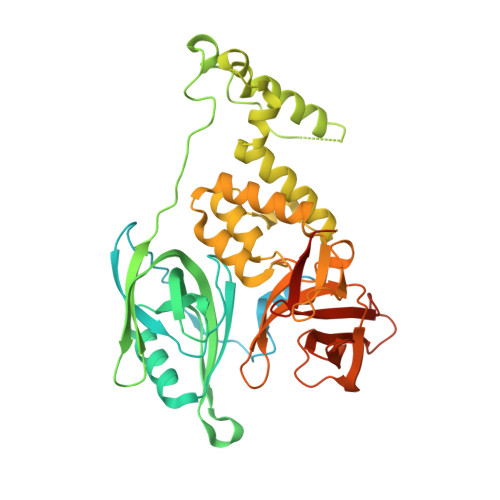
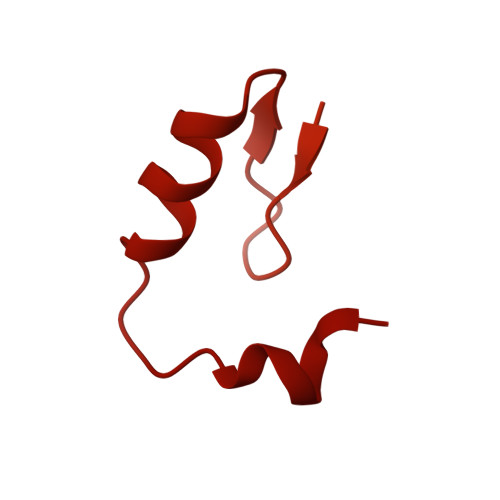
Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Mercer, J.A.M., DeCarlo, S.J., Roy Burman, S.S., Sreekanth, V., Nelson, A.T., Hunkeler, M., Chen, P.J., Donovan, K.A., Kokkonda, P., Tiwari, P.K., Shoba, V.M., Deb, A., Choudhary, A., Fischer, E.S., Liu, D.R.(2024) Science 383: eadk4422-eadk4422
- PubMed: 38484051
- DOI: https://doi.org/10.1126/science.adk4422
- Primary Citation of Related Structures:
8TNP, 8TNQ, 8TNR - PubMed Abstract:
Conditional protein degradation tags (degrons) are usually >100 amino acids long or are triggered by small molecules with substantial off-target effects, thwarting their use as specific modulators of endogenous protein levels. We developed a phage-assisted continuous evolution platform for molecular glue complexes (MG-PACE) and evolved a 36-amino acid zinc finger (ZF) degron (SD40) that binds the ubiquitin ligase substrate receptor cereblon in complex with PT-179, an orthogonal thalidomide derivative. Endogenous proteins tagged in-frame with SD40 using prime editing are degraded by otherwise inert PT-179. Cryo-electron microscopy structures of SD40 in complex with ligand-bound cereblon revealed mechanistic insights into the molecular basis of SD40's activity and specificity. Our efforts establish a system for continuous evolution of molecular glue complexes and provide ZF tags that overcome shortcomings associated with existing degrons.
- Merkin Institute of Transformative Technologies in Healthcare, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
Organizational Affiliation: