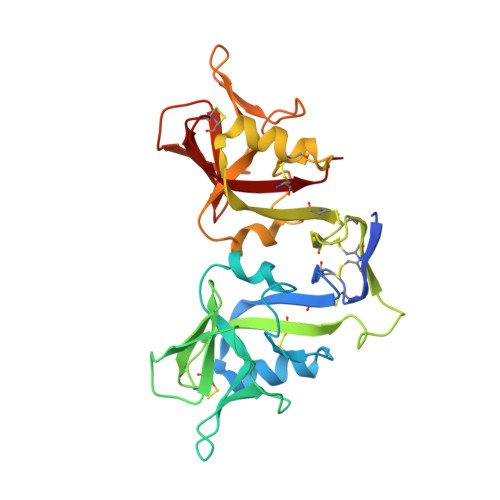
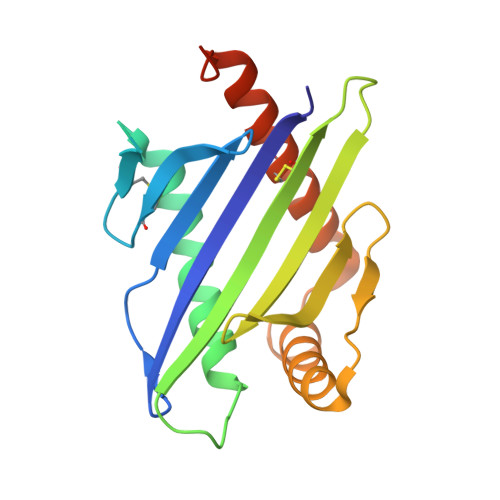
Preclinical characterization of Pan-NKG2D ligand-binding NKG2D receptor decoys.
Rupert, P.B., Buerger, M., Girard, E.J., Frutoso, M., Parrilla, D., Ng, K., Gooley, T., Groh, V., Strong, R.K.(2024) Heliyon 10: e28583-e28583
- PubMed: 38586421
- DOI: https://doi.org/10.1016/j.heliyon.2024.e28583
- Primary Citation of Related Structures:
8TLZ, 8TM0, 8TM2 - PubMed Abstract:
NKG2D and its ligands are critical regulators of protective immune responses controlling infections and cancer, defining a crucial immune signaling axis. Current therapeutic efforts targeting this axis almost exclusively aim at enhancing NKG2D-mediated effector functions. However, this axis can drive disease processes when dysregulated, in particular, driving stem-like cancer cell reprogramming and tumorigenesis through receptor/ligand self-stimulation on tumor cells. Despite complexities with its structure and biology, we developed multiple novel engineered proteins that functionally serve as axis-blocking NKG2D "decoys" and report biochemical, structural, in vitro , and in vivo evaluation of their functionality.
- Division of Basic Science, Fred Hutchinson Cancer Center, Seattle, WA, United States.
Organizational Affiliation: