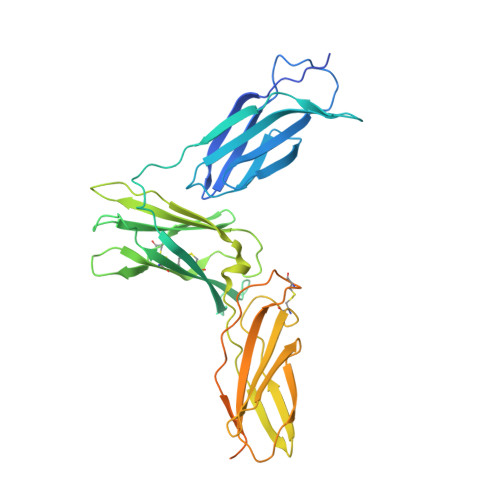
Structure of the interleukin-5 receptor complex exemplifies the organizing principle of common beta cytokine signaling.
Caveney, N.A., Rodriguez, G.E., Pollmann, C., Meyer, T., Borowska, M.T., Wilson, S.C., Wang, N., Xiang, X., Householder, K.D., Tao, P., Su, L.L., Saxton, R.A., Piehler, J., Garcia, K.C.(2024) Mol Cell 84: 1995-2005.e7
- PubMed: 38614096
- DOI: https://doi.org/10.1016/j.molcel.2024.03.023
- Primary Citation of Related Structures:
8TLD - PubMed Abstract:
Cytokines regulate immune responses by binding to cell surface receptors, including the common subunit beta (βc), which mediates signaling for GM-CSF, IL-3, and IL-5. Despite known roles in inflammation, the structural basis of IL-5 receptor activation remains unclear. We present the cryo-EM structure of the human IL-5 ternary receptor complex, revealing architectural principles for IL-5, GM-CSF, and IL-3. In mammalian cell culture, single-molecule imaging confirms hexameric IL-5 complex formation on cell surfaces. Engineered chimeric receptors show that IL-5 signaling, as well as IL-3 and GM-CSF, can occur through receptor heterodimerization, obviating the need for higher-order assemblies of βc dimers. These findings provide insights into IL-5 and βc receptor family signaling mechanisms, aiding in the development of therapies for diseases involving deranged βc signaling.
- Departments of Molecular and Cellular Physiology and Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA; Centre for Blood Research, University of British Columbia, Vancouver, BC, Canada. Electronic address: nathanael.caveney@ubc.ca.
Organizational Affiliation: