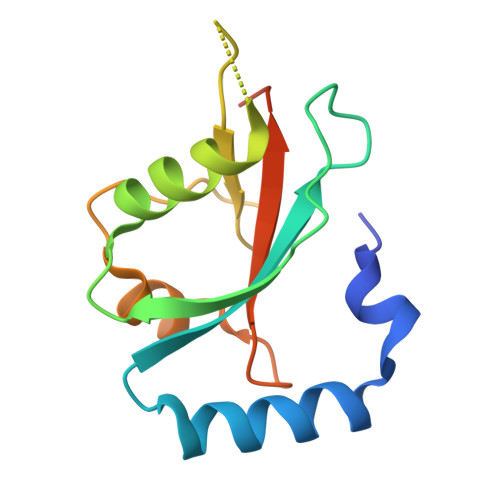
Structure of the human autophagy factor EPG5 and the molecular basis of its conserved mode of interaction with Atg8-family proteins.
Cheung, Y.W.S., Nam, S.E., Fairlie, G.M.J., Scheu, K., Bui, J.M., Shariati, H.R., Gsponer, J., Yip, C.K.(2025) Autophagy 21: 1173-1191
- PubMed: 39809444
- DOI: https://doi.org/10.1080/15548627.2024.2447213
- Primary Citation of Related Structures:
8TGF, 8TGX - PubMed Abstract:
The multi-step macroautophagy/autophagy process ends with the cargo-laden autophagosome fusing with the lysosome to deliver the materials to be degraded. The metazoan-specific autophagy factor EPG5 plays a crucial role in this step by enforcing fusion specificity and preventing mistargeting. How EPG5 exerts its critical function and how its deficiency leads to diverse phenotypes of the rare multi-system disorder Vici syndrome are not fully understood. Here, we report the first structure of human EPG5 (HsEPG5) determined by cryo-EM and AlphaFold2 modeling. Our structure revealed that HsEPG5 is constructed from helical bundles analogous to tethering factors in membrane trafficking pathways but contains a unique protruding thumb domain positioned adjacent to the atypical tandem LIR motifs involved in interaction with the GABARAP subfamily of Atg8-family proteins. Our NMR spectroscopic, molecular dynamics simulations and AlphaFold modeling studies showed that the HsEPG5 tandem LIR motifs only bind the canonical LIR docking site (LDS) on GABARAP without engaging in multivalent interaction. Our co-immunoprecipitation analysis further indicated that full-length HsEPG5-GABARAP interaction is mediated primarily by LIR1. Finally, our biochemical affinity isolation, X-ray crystallographic analysis, affinity measurement, and AlphaFold modeling demonstrated that this mode of binding is observed between Caenorhabditis elegans EPG-5 and its Atg8-family proteins LGG-1 and LGG-2. Collectively our work generated novel insights into the structural properties of EPG5 and how it potentially engages with the autophagosome to confer fusion specificity. ABBREVIATIONS : ATG: autophagy related; CSP: chemical shift perturbation; eGFP: enhanced green fluoresent protein; EM: electron microscopy; EPG5: ectopic P-granules 5 autophagy tethering factor; GST: glutathione S-transferase; HP: hydrophobic pocket; HSQC: heteronuclear single-quantum correlation; ITC: isothermal titration calorimetry; LDS: LC3 docking site; LIR: LC3-interacting region; MD: molecular dynamics; NMR: nuclear magnetic resonance; TEV: tobacco etch virus.
- Life Sciences Institute, Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: