Crystal structure of human DDX1 helicase in complex with ADP
Zeng, H., Dong, A., Li, Y., Yen, H., Hejazi, Z., Seitova, A., Arrowsmith, C.H., Edwards, A.M., Halabelian, L.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent RNA helicase DDX1 | 470 | Homo sapiens | Mutation(s): 0 Gene Names: DDX1 EC: 3.6.4.13 | 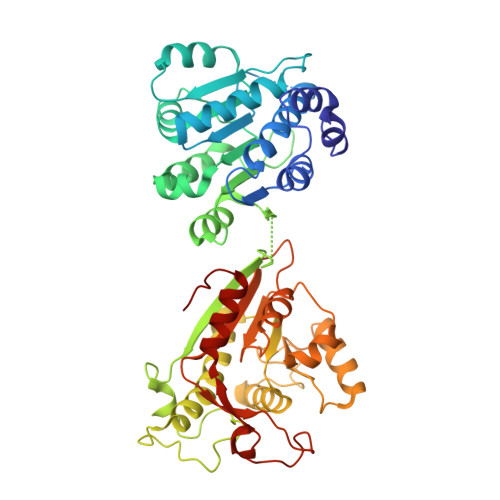 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q92499 (Homo sapiens) Explore Q92499 Go to UniProtKB: Q92499 | |||||
PHAROS: Q92499 GTEx: ENSG00000079785 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q92499 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | B [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| ZN Query on ZN | D [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | C [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.708 | α = 90 |
| b = 71.517 | β = 90 |
| c = 138.496 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-3000 | data scaling |
| PHASER | phasing |
| HKL-3000 | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute on Aging (NIH/NIA) | United States | U54 AG065187 |