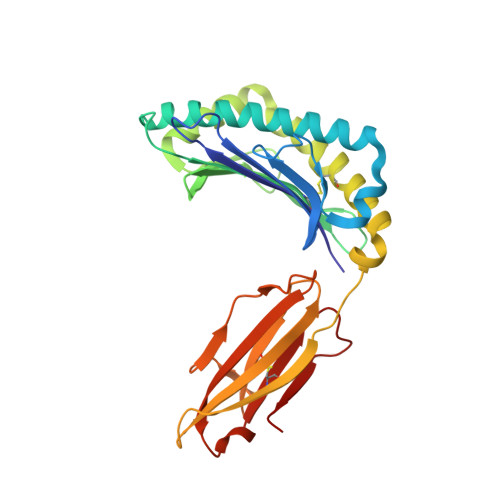
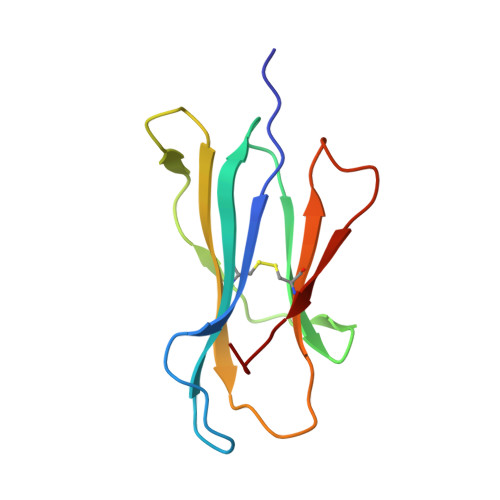
Accurate modeling of peptide-MHC structures with AlphaFold.
Mikhaylov, V., Brambley, C.A., Keller, G.L.J., Arbuiso, A.G., Weiss, L.I., Baker, B.M., Levine, A.J.(2024) Structure 32: 228
- PubMed: 38113889
- DOI: https://doi.org/10.1016/j.str.2023.11.011
- Primary Citation of Related Structures:
8TBV, 8TBW, 8U9G - PubMed Abstract:
Major histocompatibility complex (MHC) proteins present peptides on the cell surface for T cell surveillance. Reliable in silico prediction of which peptides would be presented and which T cell receptors would recognize them is an important problem in structural immunology. Here, we introduce an AlphaFold-based pipeline for predicting the three-dimensional structures of peptide-MHC complexes for class I and class II MHC molecules. Our method demonstrates high accuracy, outperforming existing tools in class I modeling accuracy and class II peptide register prediction. We validate its performance and utility with new experimental data on a recently described cancer neoantigen/wild-type peptide pair and explore applications toward improving peptide-MHC binding prediction.
- The Simons Center for Systems Biology, Institute for Advanced Study, 1 Einstein Drive, Princeton, NJ 08540, USA. Electronic address: vmikhayl@ias.edu.
Organizational Affiliation: