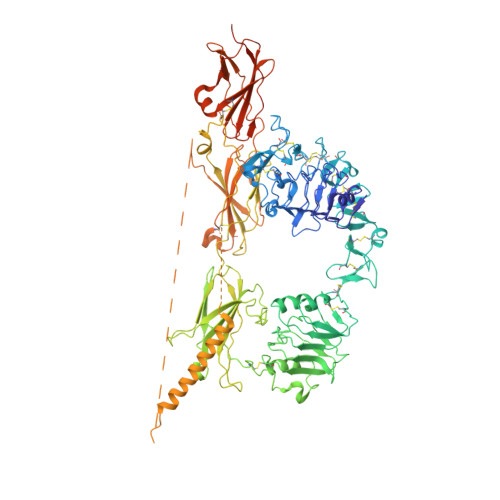
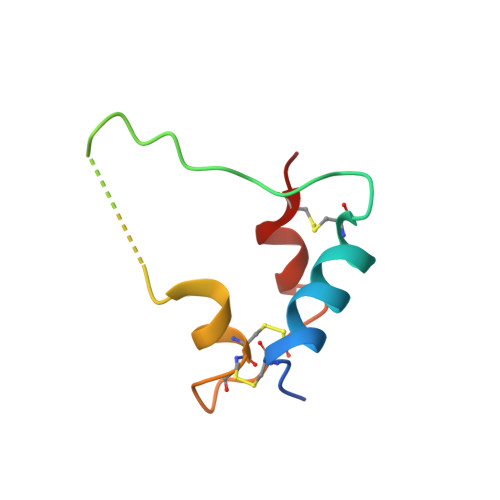
A viral insulin-like peptide inhibits IGF-1 receptor phosphorylation and regulates IGF1R gene expression.
Chrudinova, M., Kirk, N.S., Chuard, A., Venugopal, H., Zhang, F., Lubos, M., Gelfanov, V., Panikova, T., Zakova, L., Cutone, J., Mojares, M., DiMarchi, R., Jiracek, J., Altindis, E.(2024) Mol Metab 80: 101863-101863
- PubMed: 38182007
- DOI: https://doi.org/10.1016/j.molmet.2023.101863
- Primary Citation of Related Structures:
8TAN - PubMed Abstract:
The insulin/IGF superfamily is conserved across vertebrates and invertebrates. Our team has identified five viruses containing genes encoding viral insulin/IGF-1 like peptides (VILPs) closely resembling human insulin and IGF-1. This study aims to characterize the impact of Mandarin fish ranavirus (MFRV) and Lymphocystis disease virus-Sa (LCDV-Sa) VILPs on the insulin/IGF system for the first time. We chemically synthesized single chain (sc, IGF-1 like) and double chain (dc, insulin like) forms of MFRV and LCDV-Sa VILPs. Using cell lines overexpressing either human insulin receptor isoform A (IR-A), isoform B (IR-B) or IGF-1 receptor (IGF1R), and AML12 murine hepatocytes, we characterized receptor binding, insulin/IGF signaling. We further characterized the VILPs' effects of proliferation and IGF1R and IR gene expression, and compared them to native ligands. Additionally, we performed insulin tolerance test in CB57BL/6 J mice to examine in vivo effects of VILPs on blood glucose levels. Finally, we employed cryo-electron microscopy (cryoEM) to analyze the structure of scMFRV-VILP in complex with the IGF1R ectodomain. VILPs can bind to human IR and IGF1R, stimulate receptor autophosphorylation and downstream signaling pathways. Notably, scMFRV-VILP exhibited a particularly strong affinity for IGF1R, with a mere 10-fold decrease compared to human IGF-1. At high concentrations, scMFRV-VILP selectively reduced IGF-1 stimulated IGF1R autophosphorylation and Erk phosphorylation (Ras/MAPK pathway), while leaving Akt phosphorylation (PI3K/Akt pathway) unaffected, indicating a potential biased inhibitory function. Prolonged exposure to MFRV-VILP led to a significant decrease in IGF1R gene expression in IGF1R overexpressing cells and AML12 hepatocytes. Furthermore, insulin tolerance test revealed scMFRV-VILP's sustained glucose-lowering effect compared to insulin and IGF-1. Finally, cryo-EM analysis revealed that scMFRV-VILP engages with IGF1R in a manner closely resembling IGF-1 binding, resulting in a highly analogous structure. This study introduces MFRV and LCDV-Sa VILPs as novel members of the insulin/IGF superfamily. Particularly, scMFRV-VILP exhibits a biased inhibitory effect on IGF1R signaling at high concentrations, selectively inhibiting IGF-1 stimulated IGF1R autophosphorylation and Erk phosphorylation, without affecting Akt phosphorylation. In addition, MFRV-VILP specifically regulates IGF-1R gene expression and IGF1R protein levels without affecting IR. CryoEM analysis confirms that scMFRV-VILP' binding to IGF1R is mirroring the interaction pattern observed with IGF-1. These findings offer valuable insights into IGF1R action and inhibition, suggesting potential applications in development of IGF1R specific inhibitors and advancing long-lasting insulins.
- Boston College Biology Department, Chestnut Hill, MA, USA.
Organizational Affiliation: