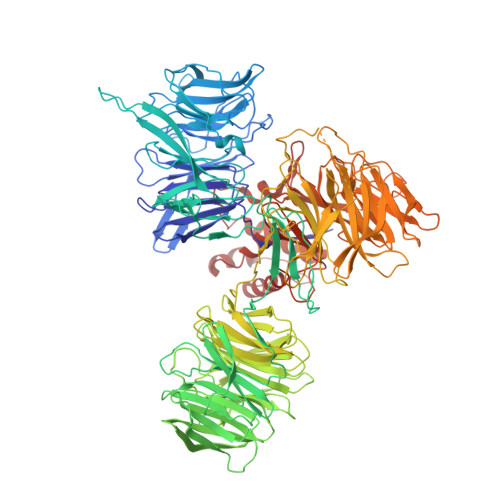
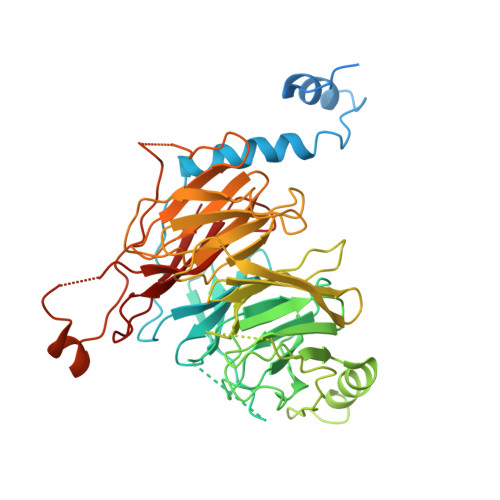
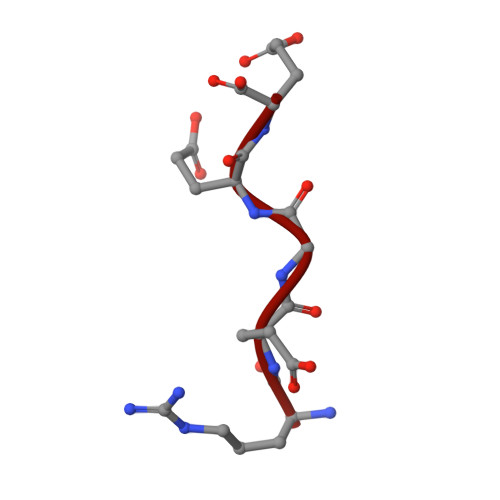
Probing the CRL4 DCAF12 interactions with MAGEA3 and CCT5 di-Glu C-terminal degrons.
Righetto, G.L., Yin, Y., Duda, D.M., Vu, V., Szewczyk, M.M., Zeng, H., Li, Y., Loppnau, P., Mei, T., Li, Y.Y., Seitova, A., Patrick, A.N., Brazeau, J.F., Chaudhry, C., Barsyte-Lovejoy, D., Santhakumar, V., Halabelian, L.(2024) PNAS Nexus 3: pgae153-pgae153
- PubMed: 38665159
- DOI: https://doi.org/10.1093/pnasnexus/pgae153
- Primary Citation of Related Structures:
8T9A - PubMed Abstract:
Damaged DNA-binding protein-1 (DDB1)- and CUL4-associated factor 12 (DCAF12) serves as the substrate recognition component within the Cullin4-RING E3 ligase (CRL4) complex, capable of identifying C-terminal double-glutamic acid degrons to promote the degradation of specific substrates through the ubiquitin proteasome system. Melanoma-associated antigen 3 (MAGEA3) and T-complex protein 1 subunit epsilon (CCT5) proteins have been identified as cellular targets of DCAF12. To further characterize the interactions between DCAF12 and both MAGEA3 and CCT5, we developed a suite of biophysical and proximity-based cellular NanoBRET assays showing that the C-terminal degron peptides of both MAGEA3 and CCT5 form nanomolar affinity interactions with DCAF12 in vitro and in cells. Furthermore, we report here the 3.17 Å cryo-EM structure of DDB1-DCAF12-MAGEA3 complex revealing the key DCAF12 residues responsible for C-terminal degron recognition and binding. Our study provides new insights and tools to enable the discovery of small molecule handles targeting the WD40-repeat domain of DCAF12 for future proteolysis targeting chimera design and development.
- Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5G 1L7, Canada.
Organizational Affiliation: