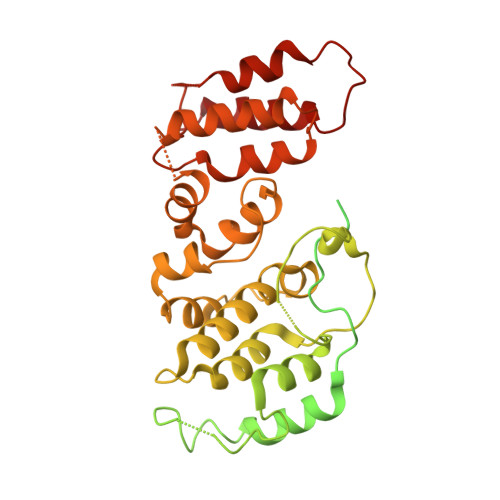
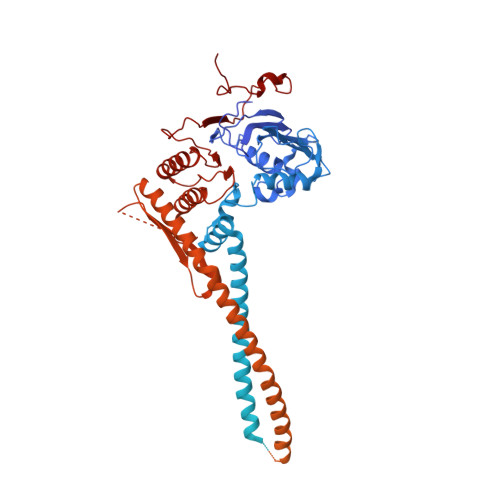
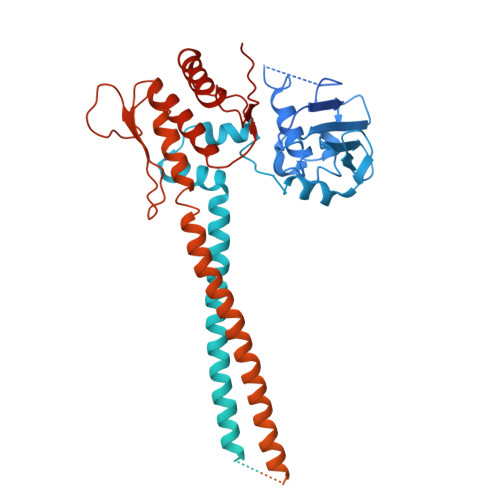
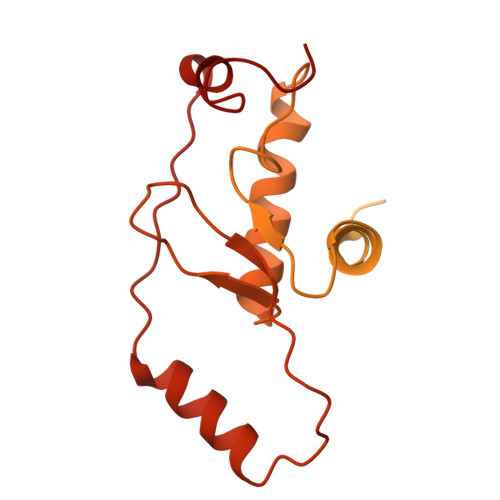
Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Li, S., Yu, Y., Zheng, J., Miller-Browne, V., Ser, Z., Kuang, H., Patel, D.J., Zhao, X.(2023) Proc Natl Acad Sci U S A 120: e2310924120-e2310924120
- PubMed: 37903273
- DOI: https://doi.org/10.1073/pnas.2310924120
- Primary Citation of Related Structures:
8T8E, 8T8F - PubMed Abstract:
The Smc5/6 complex (Smc5/6) is important for genome replication and repair in eukaryotes. Its cellular functions are closely linked to the ATPase activity of the Smc5 and Smc6 subunits. This activity requires the dimerization of the motor domains of the two SMC subunits and is regulated by the six non-SMC subunits (Nse1 to Nse6). Among the NSEs, Nse5 and Nse6 form a stable subcomplex (Nse5-6) that dampens the ATPase activity of the complex. However, the underlying mechanisms and biological significance of this regulation remain unclear. Here, we address these issues using structural and functional studies. We determined cryo-EM structures of the yeast Smc5/6 derived from complexes consisting of either all eight subunits or a subset of five subunits. Both structures reveal that Nse5-6 associates with Smc6's motor domain and the adjacent coiled-coil segment, termed the neck region. Our structural analyses reveal that this binding is compatible with motor domain dimerization but results in dislodging the Nse4 subunit from the Smc6 neck. As the Nse4-Smc6 neck interaction favors motor domain engagement and thus ATPase activity, Nse6's competition with Nse4 can explain how Nse5-6 disfavors ATPase activity. Such regulation could in principle differentially affect Smc5/6-mediated processes depending on their needs of the complex's ATPase activity. Indeed, mutagenesis data in cells provide evidence that the Nse6-Smc6 neck interaction is important for the resolution of DNA repair intermediates but not for replication termination. Our results thus provide a molecular basis for how Nse5-6 modulates the ATPase activity and cellular functions of Smc5/6.
- Molecular Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065.
Organizational Affiliation: