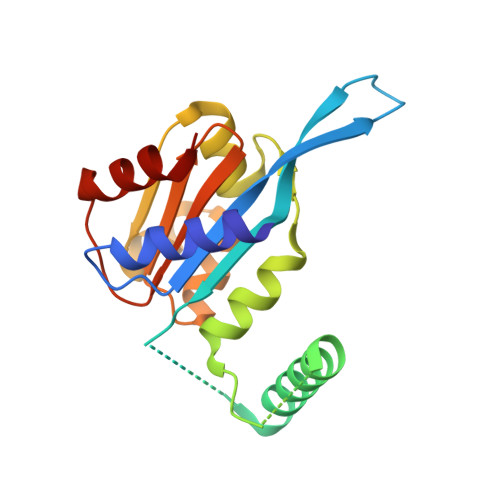
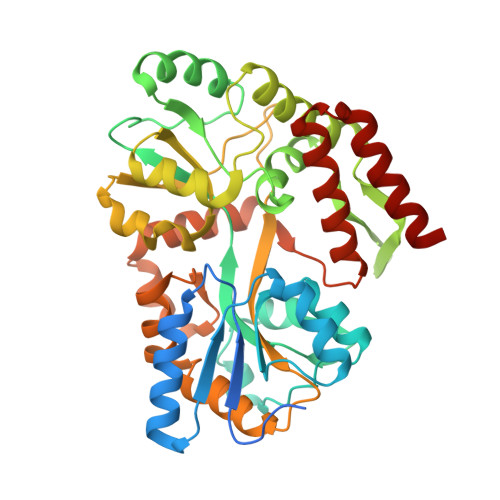
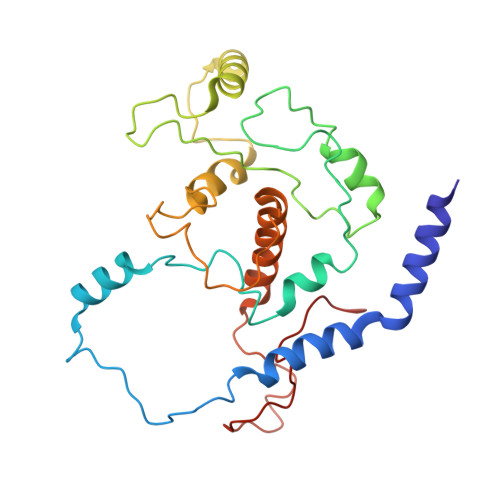
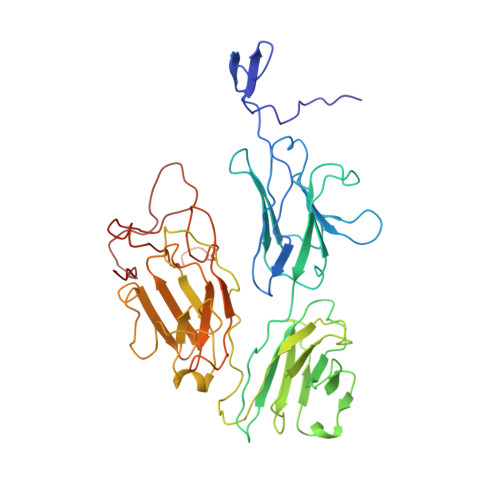
Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Zhou, X.E., Zhang, Y., Yao, J., Zheng, J., Zhou, Y., He, Q., Moreno, J., Lam, V.Q., Cao, X., Sugimoto, K., Vanegas-Cano, L., Kariapper, L., Suino-Powell, K., Zhu, Y., Novick, S., Griffin, P.R., Zhang, F., Howe, G.A., Melcher, K.(2023) Plant Commun 4: 100639-100639
- PubMed: 37322867
- DOI: https://doi.org/10.1016/j.xplc.2023.100639
- Primary Citation of Related Structures:
8T2I - PubMed Abstract:
Jasmonates (JAs) are plant hormones with crucial roles in development and stress resilience. They activate MYC transcription factors by mediating the proteolysis of MYC inhibitors called JAZ proteins. In the absence of JA, JAZ proteins bind and inhibit MYC through the assembly of MYC-JAZ-Novel Interactor of JAZ (NINJA)-TPL repressor complexes. However, JAZ and NINJA are predicted to be largely intrinsically unstructured, which has precluded their experimental structure determination. Through a combination of biochemical, mutational, and biophysical analyses and AlphaFold-derived ColabFold modeling, we characterized JAZ-JAZ and JAZ-NINJA interactions and generated models with detailed, high-confidence domain interfaces. We demonstrate that JAZ, NINJA, and MYC interface domains are dynamic in isolation and become stabilized in a stepwise order upon complex assembly. By contrast, most JAZ and NINJA regions outside of the interfaces remain highly dynamic and cannot be modeled in a single conformation. Our data indicate that the small JAZ Zinc finger expressed in Inflorescence Meristem (ZIM) motif mediates JAZ-JAZ and JAZ-NINJA interactions through separate surfaces, and our data further suggest that NINJA modulates JAZ dimerization. This study advances our understanding of JA signaling by providing insights into the dynamics, interactions, and structure of the JAZ-NINJA core of the JA repressor complex.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503, USA.
Organizational Affiliation: