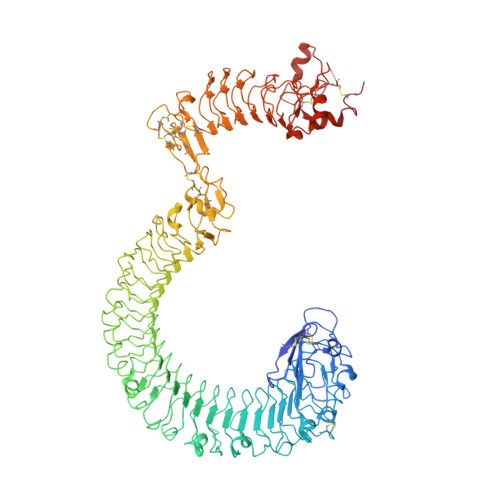
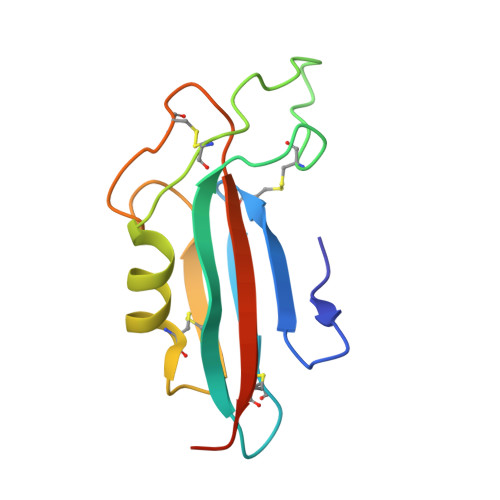
Structural basis and functional roles for Toll-like receptor binding to Latrophilin in C. elegans development.
Carmona-Rosas, G., Li, J., Smith, J.J., Nawrocka, W.I., Cheng, S., Baltrusaitis, E.E., Zhao, M., Arac, D., Kratsios, P., Ozkan, E.(2025) Nat Struct Mol Biol 32: 1683-1696
- PubMed: 40588662
- DOI: https://doi.org/10.1038/s41594-025-01592-8
- Primary Citation of Related Structures:
8SUF - PubMed Abstract:
Latrophilins are conserved adhesion-type G-protein-coupled receptors associated with embryonic defects and lethality. However, their mechanistic roles and ligands in embryogenesis remain unknown. Here, we identified TOL-1, the sole Toll-like receptor in Caenorhabditis elegans, as a ligand for the C. elegans latrophilin, LAT-1. The extracellular lectin domain of LAT-1 directly binds to the second leucine-rich repeat domain of TOL-1. The crystal structure and cryo-electron microscopy density map of the LAT-1-TOL-1 extracellular region complex reveal a one-to-one lectin domain interaction with the convex face of a leucine-rich repeat domain. In C. elegans, endogenous mRNA and protein localization analyses showed mutually exclusive sites of expression, suggesting that in vivo LAT-1-TOL-1 interactions mostly occur in trans. Mutagenesis of key interface residues that disrupt the LAT-1-TOL-1 interaction led to partial lethality and malformed embryos. Thus, TOL-1 binding to LAT-1 represents a receptor-ligand axis essential for animal development.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, USA.
Organizational Affiliation: