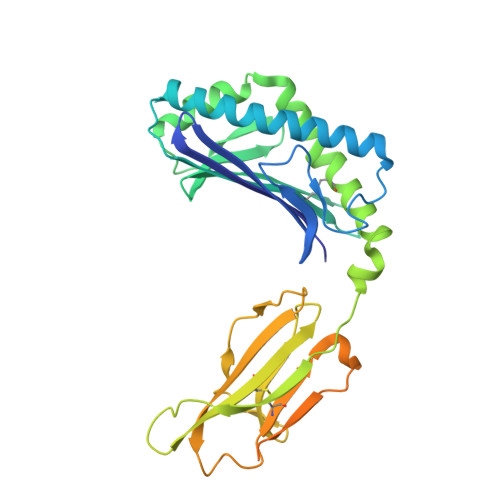
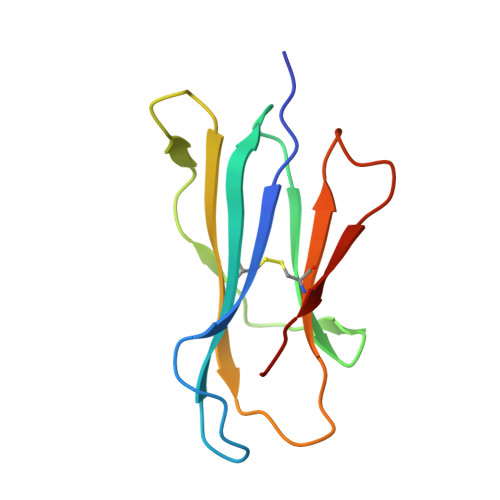
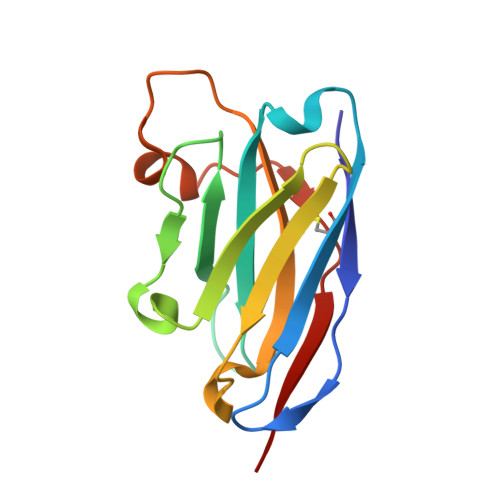
Enhanced CD1d phosphatidylserine presentation using a single-domain antibody promotes immunomodulatory CD1d-TIM-3 interactions.
Lameris, R., Shahine, A., Veth, M., Westerman, B., Godfrey, D.I., Lutje Hulsik, D., Brouwer, P., Rossjohn, J., de Gruijl, T.D., van der Vliet, H.J.(2023) J Immunother Cancer 11
- PubMed: 38040419
- DOI: https://doi.org/10.1136/jitc-2023-007631
- Primary Citation of Related Structures:
8SOS - PubMed Abstract:
CD1d is a monomorphic major histocompatibility complex class I-like molecule that presents lipid antigens to distinct T-cell subsets and can be expressed by various malignancies. Antibody-mediated targeting of CD1d on multiple myeloma cells was reported to induce apoptosis and could therefore constitute a novel therapeutic approach. To determine how a CD1d-specific single-domain antibody (VHH) enhances binding of the early apoptosis marker annexin V to CD1d + tumor cells we use in vitro cell-based assays and CRISPR-Cas9-mediated gene editing, and to determine the structure of the VHH1D17-CD1d(endogenous lipid) complex we use X-ray crystallography. Anti-CD1d VHH1D17 strongly enhances annexin V binding to CD1d + tumor cells but this does not reflect induction of apoptosis. Instead, we show that VHH1D17 enhances presentation of phosphatidylserine (PS) in CD1d and that this is saposin dependent. The crystal structure of the VHH1D17-CD1d(endogenous lipid) complex demonstrates that VHH1D17 binds the A'-pocket of CD1d, leaving the lipid headgroup solvent exposed, and has an electro-negatively charged patch which could be involved in the enhanced PS presentation by CD1d. Presentation of PS in CD1d does not trigger phagocytosis but leads to greatly enhanced binding of T-cell immunoglobulin and mucin domain containing molecules (TIM)-1 to TIM-3, TIM-4 and induces TIM-3 signaling. Our findings reveal the existence of an immune modulatory CD1d(PS)-TIM axis with potentially unexpected implications for immune regulation in both physiological and pathological conditions.
- Department of Medical Oncology, Cancer Center Amsterdam, Amsterdam UMC Location VUmc, Amsterdam, The Netherlands.
Organizational Affiliation: