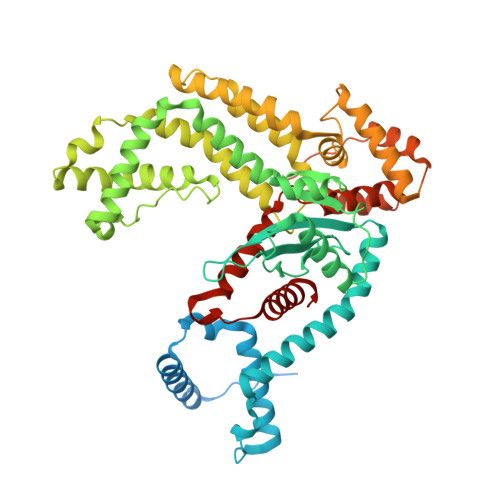
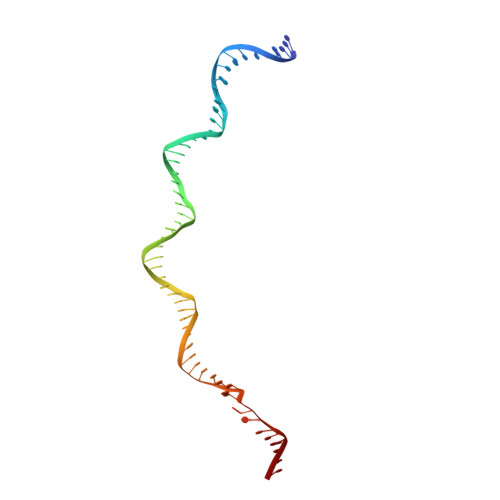
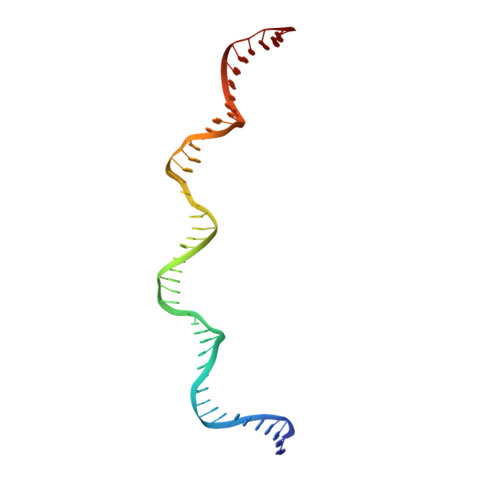
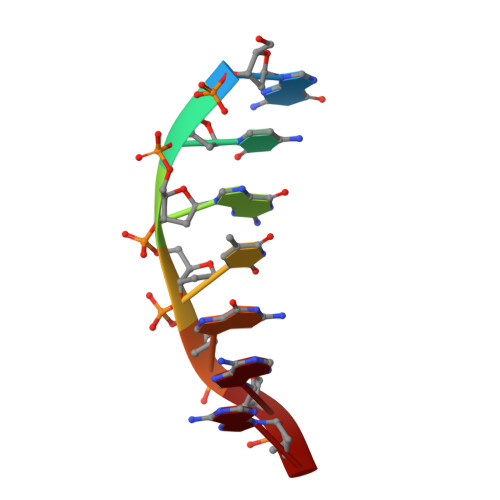
Zinc-finger BED domains drive the formation of the active Hermes transpososome by asymmetric DNA binding.
Lannes, L., Furman, C.M., Hickman, A.B., Dyda, F.(2023) Nat Commun 14: 4470-4470
- PubMed: 37491363
- DOI: https://doi.org/10.1038/s41467-023-40210-3
- Primary Citation of Related Structures:
8EB5, 8EDG, 8SJD - PubMed Abstract:
The Hermes DNA transposon is a member of the eukaryotic hAT superfamily, and its transposase forms a ring-shaped tetramer of dimers. Our investigation, combining biochemical, crystallography and cryo-electron microscopy, and in-cell assays, shows that the full-length Hermes octamer extensively interacts with its transposon left-end through multiple BED domains of three Hermes protomers contributed by three dimers explaining the role of the unusual higher-order assembly. By contrast, the right-end is bound to no BED domains at all. Thus, this work supports a model in which Hermes multimerizes to gather enough BED domains to find its left-end among the abundant genomic DNA, facilitating the subsequent interaction with the right-end.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, 20892, USA.
Organizational Affiliation: