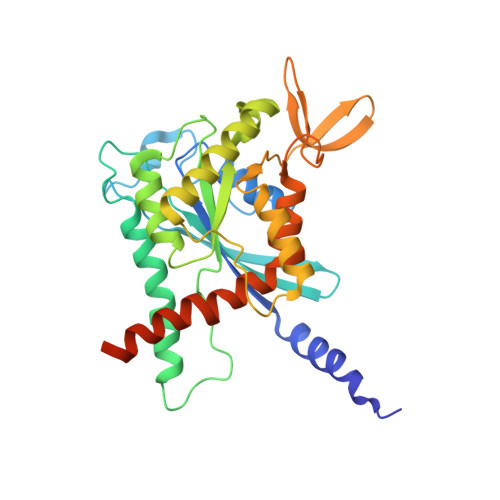
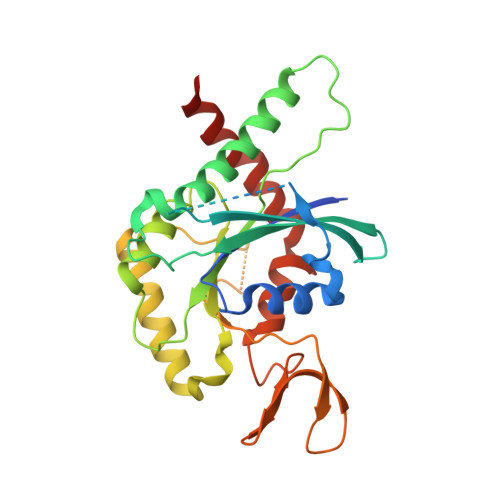
A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
Marques da Silva, R., Christe Dos Reis Saladino, G., Antonio Leonardo, D., D'Muniz Pereira, H., Andrea Sculaccio, S., Paula Ulian Araujo, A., Charles Garratt, R.(2023) J Struct Biol 215: 107983-107983
- PubMed: 37315820
- DOI: https://doi.org/10.1016/j.jsb.2023.107983
- Primary Citation of Related Structures:
8FWP, 8SGD - PubMed Abstract:
Septins, often described as the fourth component of the cytoskeleton, are structural proteins found in a vast variety of living beings. They are related to small GTPases and thus, generally, present GTPase activity which may play an important (although incompletely understood) role in their organization and function. Septins polymerize into long non-polar filaments, in which each subunit interacts with two others by alternating interfaces, NC and G. In Saccharomyces cerevisiae four septins are organized in the following manner, [Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11] n in order to form filaments. Although septins were originally discovered in yeast and much is known regarding their biochemistry and function, only limited structural information about them is currently available. Here we present crystal structures of Cdc3/Cdc10 which provide the first view of the physiological interfaces formed by yeast septins. The G-interface has properties which place it in between that formed by SEPT2/SEPT6 and SEPT7/SEPT3 in human filaments. Switch I from Cdc10 contributes significantly to the interface, whereas in Cdc3 it is largely disorded. However, the significant negative charge density of the latter suggests it may have a unique role. At the NC-interface, we describe an elegant means by which the sidechain of a glutamine from helix α 0 imitates a peptide group in order to retain hydrogen-bond continuity at the kink between helices α 5 and α 6 in the neighbouring subunit, thereby justifying the conservation of the helical distortion. Its absence from Cdc11, along with this structure's other unusual features are critically discussed by comparison with Cdc3 and Cdc10.
- Instituto de Física de São Carlos, Universidade de São Paulo, Avenida João Dagnone 1100, São Carlos, SP 13563-723, Brazil.
Organizational Affiliation: