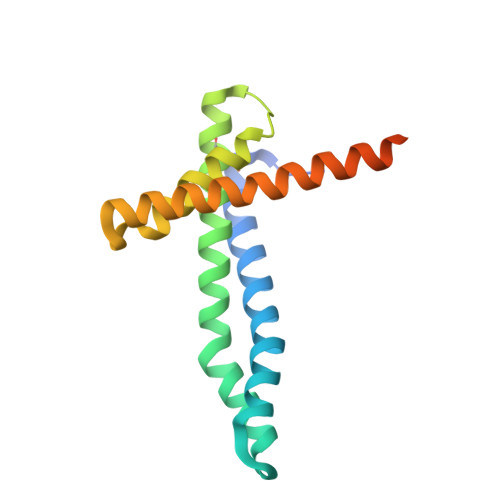
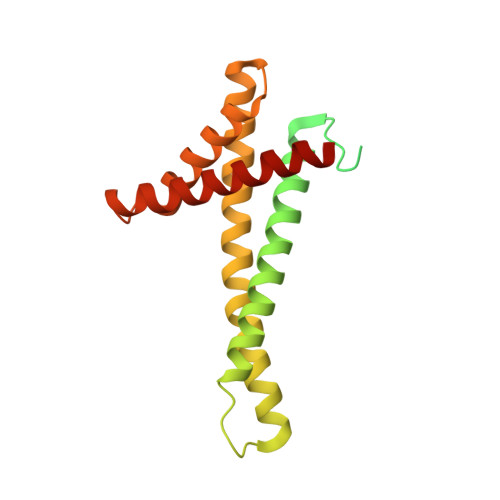
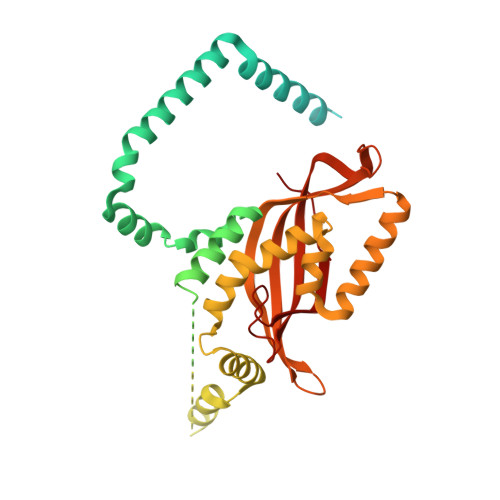
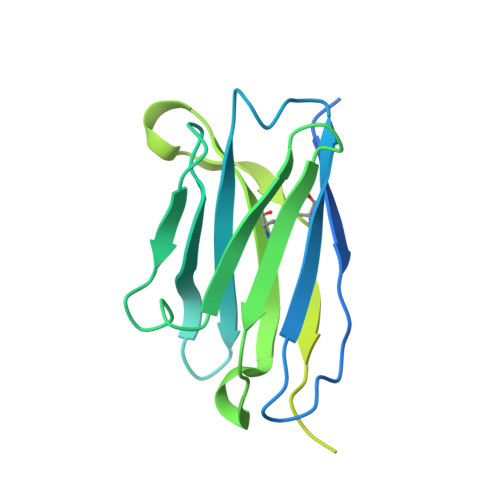
Structural basis of mitochondrial protein import by the TIM23 complex.
Sim, S.I., Chen, Y., Lynch, D.L., Gumbart, J.C., Park, E.(2023) Nature 621: 620-626
- PubMed: 37344598
- DOI: https://doi.org/10.1038/s41586-023-06239-6
- Primary Citation of Related Structures:
8E1M, 8SCX - PubMed Abstract:
Mitochondria import nearly all of their approximately 1,000-2,000 constituent proteins from the cytosol across their double-membrane envelope 1-5 . Genetic and biochemical studies have shown that the conserved protein translocase, termed the TIM23 complex, mediates import of presequence-containing proteins (preproteins) into the mitochondrial matrix and inner membrane. Among about ten different subunits of the TIM23 complex, the essential multipass membrane protein Tim23, together with the evolutionarily related protein Tim17, has long been postulated to form a protein-conducting channel 6-11 . However, the mechanism by which these subunits form a translocation path in the membrane and enable the import process remains unclear due to a lack of structural information. Here we determined the cryo-electron microscopy structure of the core TIM23 complex (heterotrimeric Tim17-Tim23-Tim44) from Saccharomyces cerevisiae. Contrary to the prevailing model, Tim23 and Tim17 themselves do not form a water-filled channel, but instead have separate, lipid-exposed concave cavities that face in opposite directions. Our structural and biochemical analyses show that the cavity of Tim17, but not Tim23, forms the protein translocation path, whereas Tim23 probably has a structural role. The results further suggest that, during translocation of substrate polypeptides, the nonessential subunit Mgr2 seals the lateral opening of the Tim17 cavity to facilitate the translocation process. We propose a new model for the TIM23-mediated protein import and sorting mechanism, a central pathway in mitochondrial biogenesis.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA.
Organizational Affiliation: