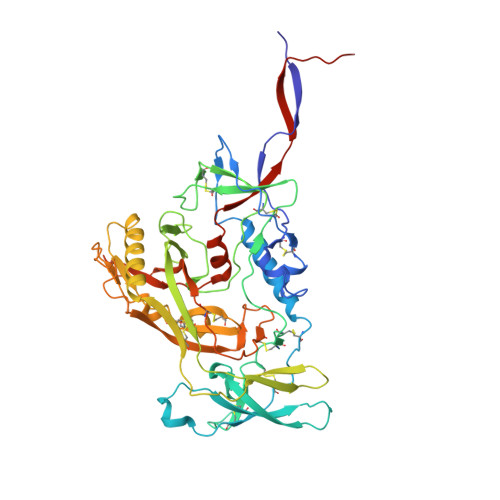
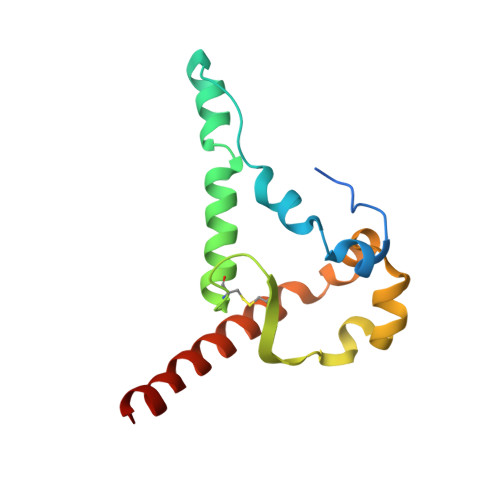
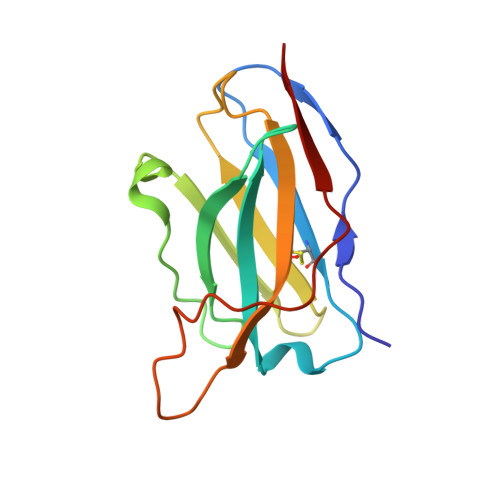
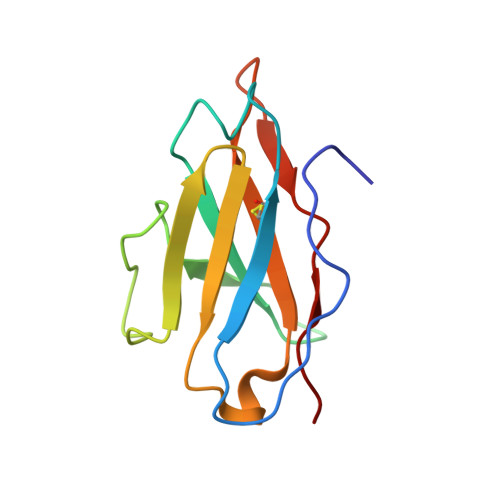
Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Henderson, R., Zhou, Y., Stalls, V., Wiehe, K., Saunders, K.O., Wagh, K., Anasti, K., Barr, M., Parks, R., Alam, S.M., Korber, B., Haynes, B.F., Bartesaghi, A., Acharya, P.(2023) Nat Commun 14: 2782-2782
- PubMed: 37188681
- DOI: https://doi.org/10.1038/s41467-023-38108-1
- Primary Citation of Related Structures:
8SAL, 8SAN, 8SAQ, 8SAR, 8SAS, 8SAT, 8SAU, 8SAV, 8SAW, 8SAX, 8SAY, 8SAZ, 8SB0, 8SB1, 8SB2, 8SB3, 8SB4, 8SB5 - PubMed Abstract:
Antibody affinity maturation enables adaptive immune responses to a wide range of pathogens. In some individuals broadly neutralizing antibodies develop to recognize rapidly mutating pathogens with extensive sequence diversity. Vaccine design for pathogens such as HIV-1 and influenza has therefore focused on recapitulating the natural affinity maturation process. Here, we determine structures of antibodies in complex with HIV-1 Envelope for all observed members and ancestral states of the broadly neutralizing HIV-1 V3-glycan targeting DH270 antibody clonal B cell lineage. These structures track the development of neutralization breadth from the unmutated common ancestor and define affinity maturation at high spatial resolution. By elucidating contacts mediated by key mutations at different stages of antibody development we identified sites on the epitope-paratope interface that are the focus of affinity optimization. Thus, our results identify bottlenecks on the path to natural affinity maturation and reveal solutions for these that will inform immunogen design aimed at eliciting a broadly neutralizing immune response by vaccination.
- Department of Medicine and Immunology, Duke University School of Medicine, Durham, NC, USA. rory.henderson@duke.edu.
Organizational Affiliation: